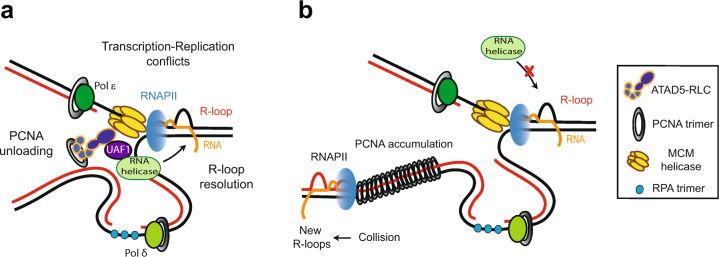
Fig. 2. Graphical model for R-loop regulation by ATAD5-RLC and ATAD5/UAF1-interacting DNA/RNA helicases.
a During normal replication, ATAD5-RLC and ATAD5/UAF1-interacting DNA/RNA helicases migrate with a replication fork. ATAD5/UAF1-interacting DNA/RNA helicases resolve R-loops ahead of the replication fork and facilitate replication fork progression. Upon replication stress, which increases transcription–replication conflicts and unscheduled R-loop formation, additional DNA/RNA helicases are recruited to the replication fork, which resolves R-loops to ensure faithful replication fork progression. Under both normal and replication stress conditions, the recruitment of helicases at the replication fork is dependent on the ATAD5/UAF1 interaction. b In ATAD5-depleted cells, reduced R-loop resolution by ATAD5/UAF1-interacting DNA/RNA helicases leads to defects in replication fork progression. In addition, PCNA and its interacting proteins accumulated on lagging strand DNA behind the forks collide with transcription machinery, which consequently increases R-loop formation at the collision site.