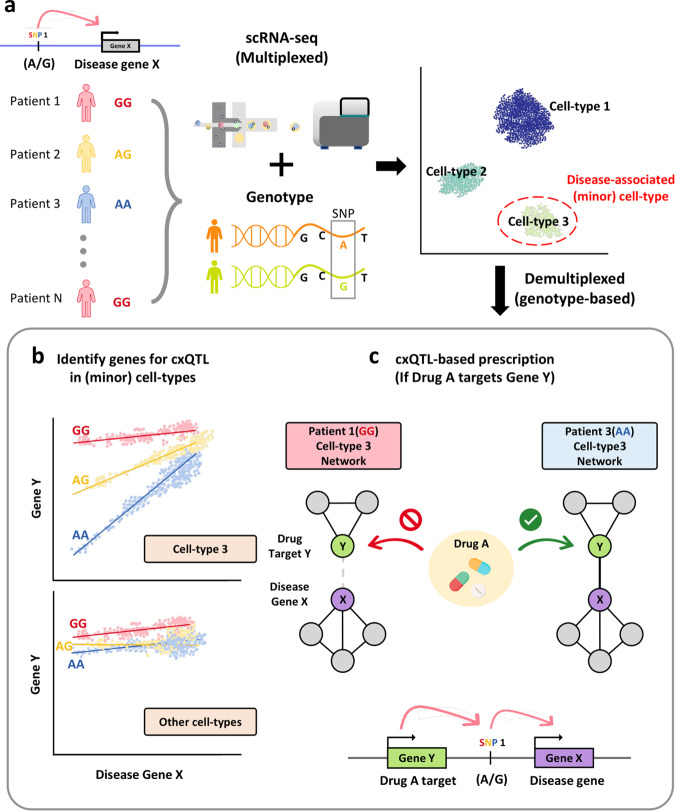
Fig. 4. Hypothesis generation from genotype-network association in single-cell network biology.
a Many disease-associated single nucleotide polymorphisms (SNPs), which are called expression QTLs (eQTLs), exert phenotypic effects through the regulation of gene expression in a cell type-specific manner. Therefore, eQTL analysis needs to be conducted for specific cell types, particularly for minor cell types. The recently developed multiplexed scRNA-seq technology along with demultiplexing based on genotype information will facilitate cell-type-specific eQTL mapping. b Some eQTL effects are dependent on the expression of other genes. This dependency is detected by genotype-specific coexpression, called coexpression QTL (cxQTL). Here, a disease gene X is coexpressed with gene Y only if its eQTL has a homozygous major allele (AA). c If the gene Y is a target of drug A that eventually inhibits the activity of disease gene X via interaction with gene Y, the genotype-dependent coregulatory interaction between genes X and Y is critical for drug action. Then, for prescription of drug A, the cxQTL genotype information can be utilized for precision medicine (e.g., prescribing it only for patients with SNP AA).