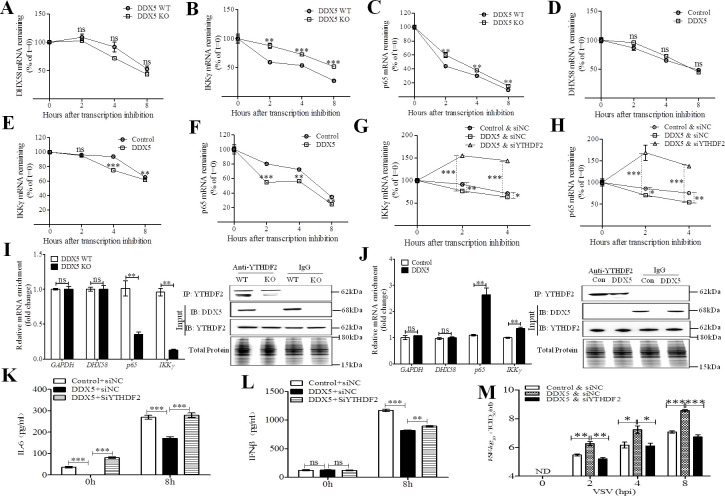
Fig 6. DDX5 inhibited innate immunity through promoting RNA decay of antiviral transcripts in a YTHDF2-dependent manner.
(A-C): DHX58, IKKγ, and p65 mRNA degradation in VSV infected DDX5-knockout (KO) MEFs treated with actinomycin D at indicated times (n = 3), the DDX5 wild type (WT) MEFs as the control. Residual RNAs were normalized to 0 h. (D-F): DHX58, IKKγ, and p65 mRNA degradation in VSV infected DDX5-expressing MEFs treated with actinomycin D at indicated times (n = 3), the DDX5 wild type (WT) MEFs as the control. Residual RNAs were normalized to 0 h. (G, H): IKKγ and p65 mRNA degradation in indicated YTHDF2-silenced MEFs treated with actinomycin-D (n = 3). Residual RNAs were normalized to 0 h. MEFs were transfected with YTHDF2 siRNA (siNC) for 24h, transfected with DDX5 (control vector) for 24h, infected with VSV, and then treated with actinomycin D at indicated times (n = 3). Residual RNAs were normalized to 0 h. (I, J): The binding of YTHDF2 and transcripts was detected via YTHDF2 RIP qRT-PCR in DDX5 KO (I) or DDX5-expressing (J) MEFs after VSV infection. DDX5 KO MEFs were infected with VSV for 6 h (I), or MEFs were transfected with DDX5 expression plasmid (DDX5) or control vector (Con) for 24 h, then infected with VSV for 6 h (J), next all the treated MEFs were subjected to YTHDF2 RIP qRT-PCR to detect GAPDH, DHX58, IKKγ, and p65. Results are presented relative to those obtained with NC or control groups, and the expression of all the indicated proteins was analyzed by western blotting. (K, L): ELISA of IL-6 or IFN-β in supernatants of YTHDF2-knockdown coupled with DDX5-expressing MEFs infected with VSV for 0 or 8 h. (M): TCID50 of VSV in YTHDF2-knockdown coupled with DDX5-expressing MEFs infected with VSV for 0, 2, 4, and 8 h. All data are mean ± SEM of biologically independent samples. n = number of biological replicates. Data are representative of three independent experiments. ND, not detected. ns, no significant difference. *p<0.05, **p<0.01, and ***p<0.001 (Student’s t-test).