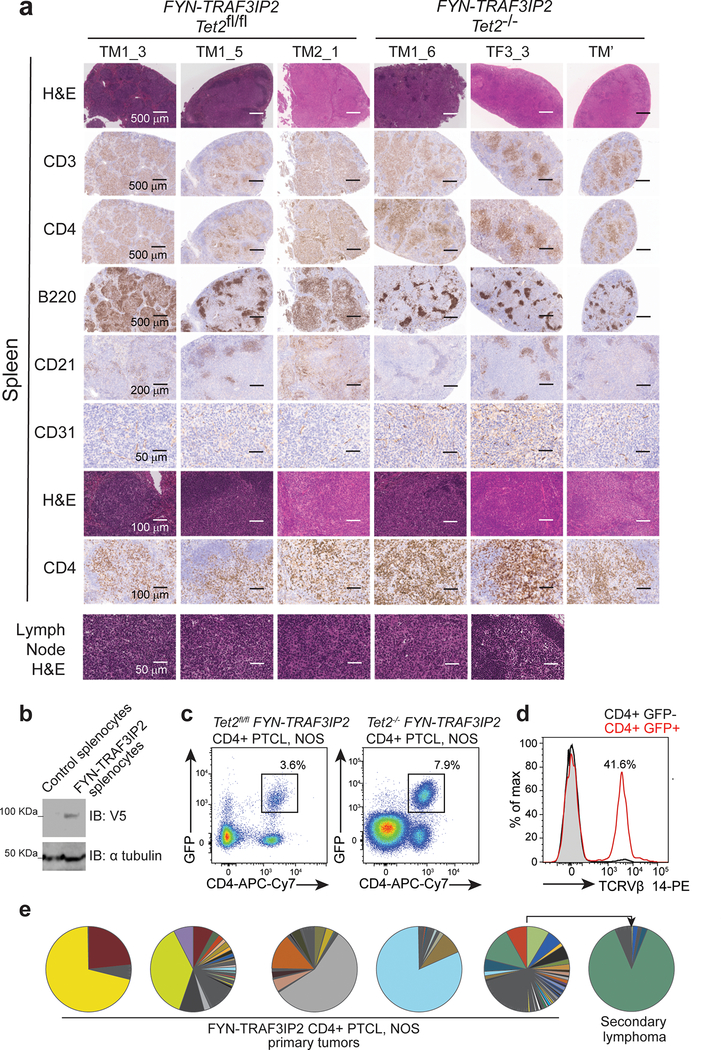
Figure 6. Histological and clonality features of FYN-TRAF3IP2-induced PTCL, NOS lymphomas.
a. Representative histopathologic micrographs of hematoxylin-eosin stained and immunohistochemistry analysis of T cells (CD3, CD4), B cells (B220), follicular dendritic cells (CD21) and vasculature (CD31) of spleen and lymph node of individual FYN-TRAF3IP2 Tet2fl/fl or Tet2−/− FYN-TRAF3IP2-induced lymphoma-bearing animals (n=3 in each group). Similar fields on consecutive sections are shown in rows 1–4 and rows 7–8. Scale bar and size are shown in the micrographs. b, Immunoblot analysis of FYN-TRAF3IP2-V5 protein expression in control splenocytes and in the lymphoma-infiltrated spleen of a FYN-TRAF3IP2 diseased mouse. c, Representative FACS plot showing GFP+ CD4+ lymphoma cells in lymphoma-infiltrated spleens from FYN-TRAF3IP2 diseased mice. d, Tcr Vβ clonality in FYN-TRAF3IP2 lymphoma cells (GFP+ CD4+) and normal CD4 T cells (GFP− CD4+) from a diseased mouse. e, Tcrb clonal distribution in FYN-TRAF3IP2-induced CD4+ primary tumors and in a representative allografted secondary lymphoma. Sectors represent the fraction of reads corresponding to individual Tcrb sequences.