Fig. 2. P. vivax-like strains are structured in two distinct clades that form a sister monophyletic lineage to the human P. vivax.
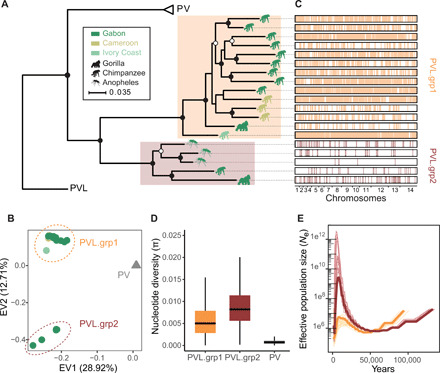
(A) ML phylogenetic tree illustrating the relationships between P. vivax (PV) and P. vivax-like (PVL) strains, rooted using P. cynomolgi (PC, M strain). Two distinct P. vivax-like groups were identified: PVL.grp1 (in orange) and PVL.grp2 (in brown). The animal pictograms on each leaf indicate the primate host, gorilla, chimpanzee, or mosquito (unknown primate host), colored according to the country of origin (Gabon, Cameroon, and Ivory Coast). Open and closed circles indicate nodes with >80 and >90% bootstrap support, respectively. (B) PCA displaying the two first eigen vectors (EVs) and the proportion of genetic variance they explain. (C) Genome-wide visualization of recent recombination events between P. vivax-like individuals. The horizontal black rectangles represent the 19 P. vivax-like genome sequences, and the vertical colored lines represent the recombining genomic segments on the recipient individual. Colors indicate the lineage membership of the donor individual. (D) Differences in nucleotide diversity (π) between P. vivax-like lineages and P. vivax strains. (E) Multiple sequentially Markovian coalescent (MSMC) estimates of the effective population size (Ne) in the two P. vivax-like groups (PVL.grp1 in orange and PVL.grp2 in brown). Lines in lighter color represent 50 bootstrap resampling replicates of randomly sampled segregating sites.
