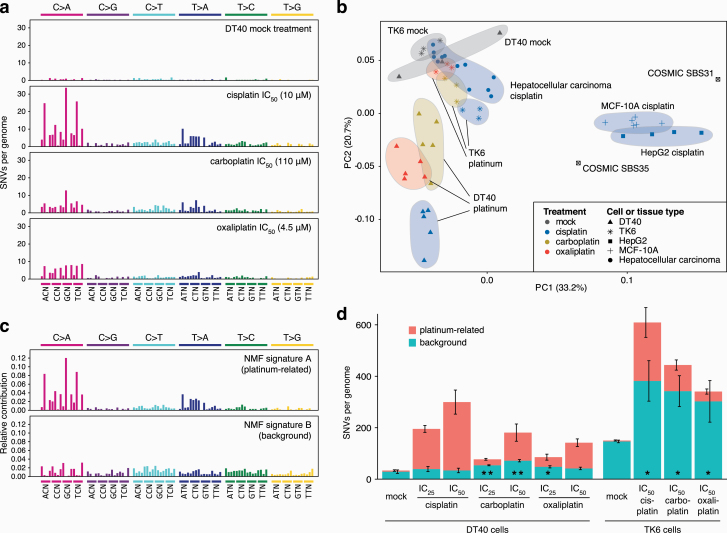
Figure 5.
Analysis of SNV mutation spectra. (a) Triplet mutation spectra of the mock, IC50 (10 µM) cisplatin, (110 µM) carboplatin and (4.5 µM) oxaliplatin treatments of DT40 cells. The middle base of each triplet, listed at the bottom, mutated as indicated at the top of the panel. The third base of the triplets, not shown for lack of space, is alphabetical. (b) Principle component analysis of normalised triplet SNV spectra from all sampled sequenced in this study, as well as from cisplatin-treated MCF-10A and HepG2 cells (35) and of 12 cisplatin-treated hepatocellular carcinoma samples. The symbols indicate cell or tissue types, the colours specify platinum drug treatments. COSMIC signatures SBS31 and SBS35 were also included and are shown as labelled. (c, d) De novo NMF of SNV mutation spectra of all sequenced samples identified two components; triplet mutation spectra of NMF signature A (platinum-related) and NMF signature B (background) are shown as the contribution of each triplet mutation type. (d) Mean SNV counts for all sequenced DT40 and in TK6 genomes, split using NMF into ‘platinum-related’ and ‘background’ signatures and then averaged by cell type and treatment as indicated. The error bars indicate the SEM of the column below, based on five DT40 or three TK6 independent treated single-cell clones. Significant differences in the mean number of mutations attributed to the ‘background’ signature, as compared to the mock treatment, are indicated at the bottom of the columns (unpaired t-test, *P < 0.05, **P < 0.01).