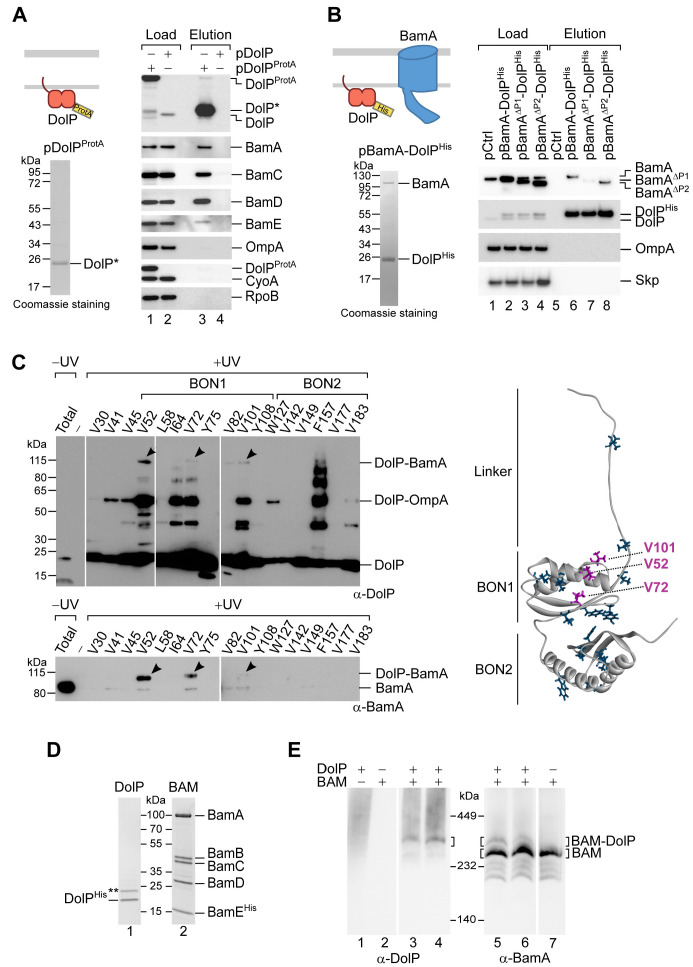
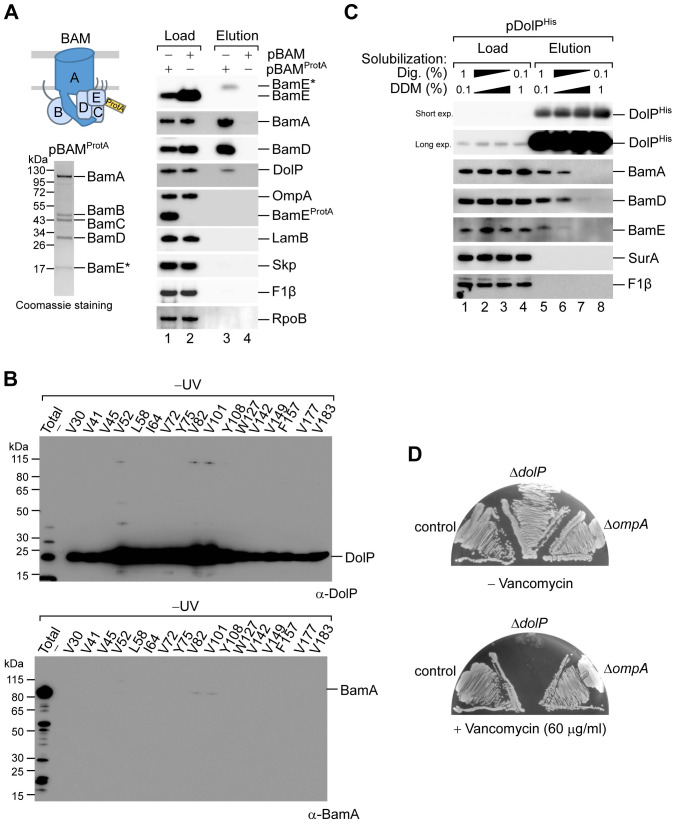
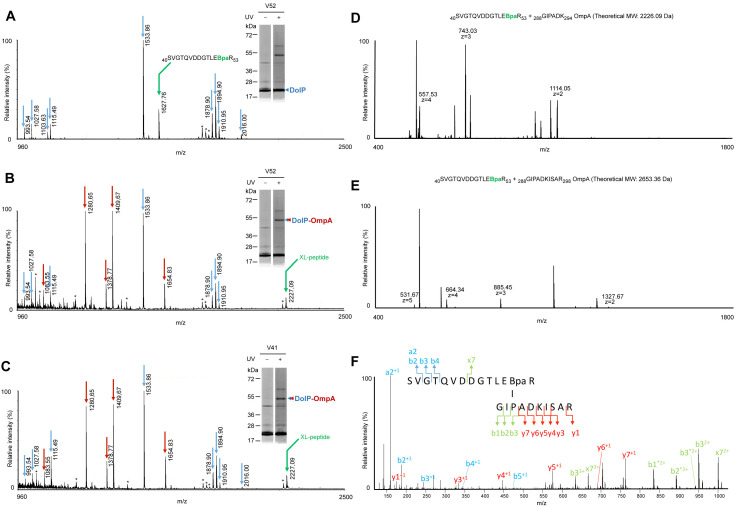
Figure 4. DolP associates with the BAM complex via an interaction with BamA.
(A) The envelope fractions of BW25113 cells carrying the indicated plasmids were solubilized with 1% (w/v) digitonin and 0.1% (w/v) DDM and subjected to IgG affinity purification of protein A-tagged DolP. The load and elution fractions were analysed by SDS-PAGE. The coomassie staining of the elution of protein A-tagged DolP is shown below the diagrams representing the overproduced protein. Blotted proteins from load and elution fractions were detected by immunolabelling using the indicated antisera. Load 0.5%; Elution 100%. The asterisk indicates the TEV-digestion product of DolPProtA. (B) The envelope fractions of BW25113 cells carrying the plasmids overproducing His-tagged DolP and the indicated BamA protein variants (deleted of POTRA1 or of POTRA2) were solubilized with 1% (w/v) digitonin and 0.1% (w/v) DDM and subjected to Ni-affinity purification. The load and elution fractions were analysed by SDS-PAGE. The coomassie staining of the elution of His-tagged DolP overproduced together with wild-type BamA is shown below the diagram representing the overproduced proteins. Blotted protein from load and elution fractions were detected by immunolabelling using the indicated antisera. Load 2%; Elution 100%. The amount of BamA co-isolated with DolPHis was normalized to the amount of BamA detected in the load fraction. The value obtained for the pBamA-DolPHis sample was set to 100%. The average of the relative amounts of co-isolated BamAΔP1 and BamAΔP2 are as follows: BamAΔP1, 16.5% (N = 2; 1st exp. 23.6%; 2nd exp. 9.3%); BamAΔP2, 81.2% (N = 2; 1st exp. 101.8%; 2nd exp. 60.6%). (C) UV photo-crosslinking of ΔdolP cells transformed with pEVOL-pBpF and pBamA-DolPHis harbouring an amber codon at the indicated position of the dolP ORF. Upon Ni-affinity chromatography of DolPHis, eluates obtained from UV irradiated samples were separated by SDS-PAGE and analysed by immunoblotting using the indicated antisera. The total envelope fraction of cells expressing DolPHis with Bpa at position V52 (non-irradiated) is shown in the first lane and serves as a reference for the migration of non-crosslinked DolP and BamA. Arrowheads indicate crosslinked products detected with both DolP and BamA antisera. Analysis of eluates obtained from non-irradiated samples are shown in Figure 4—figure supplement 1B. The amino acid residues replaced with Bpa are indicated on the structure of DolP, PDB: 7A2D (Bryant et al., 2020). In purple are the positions crosslinked to BamA. (D) The envelope fraction of BW25113 cells overproducing DolPHis or the BAM complex containing C-terminally His-tagged BamE was subjected to protein extraction with 1% (w/v) DDM, Ni-affinity purification, and gel filtration chromatography. The elution fractions were analysed by SDS-PAGE and coomassie staining. The double asterisk indicates a contaminant protein in the elution of DolP. (E) Roughly equimolar quantities of purified His-tagged BAM complex and DolP were incubated alone for 1 hr at 4°C (lanes 1, 2, and 7), or together for 1 hr at 4°C (lanes 3 and 6) or for 30 min at 25°C (lanes 4 and 5), prior to blue native-PAGE and immunoblotting using the indicated antisera.