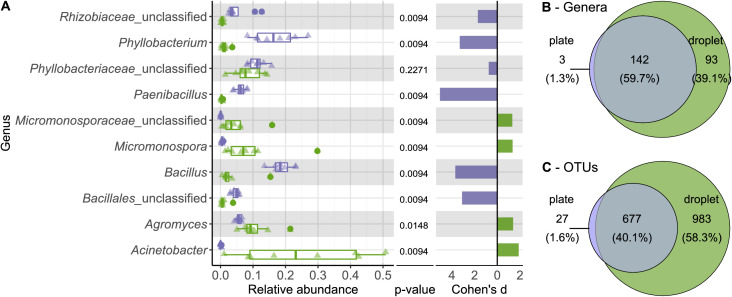
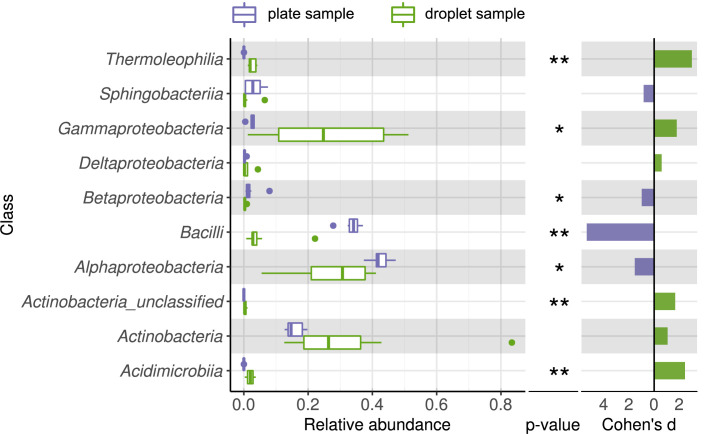
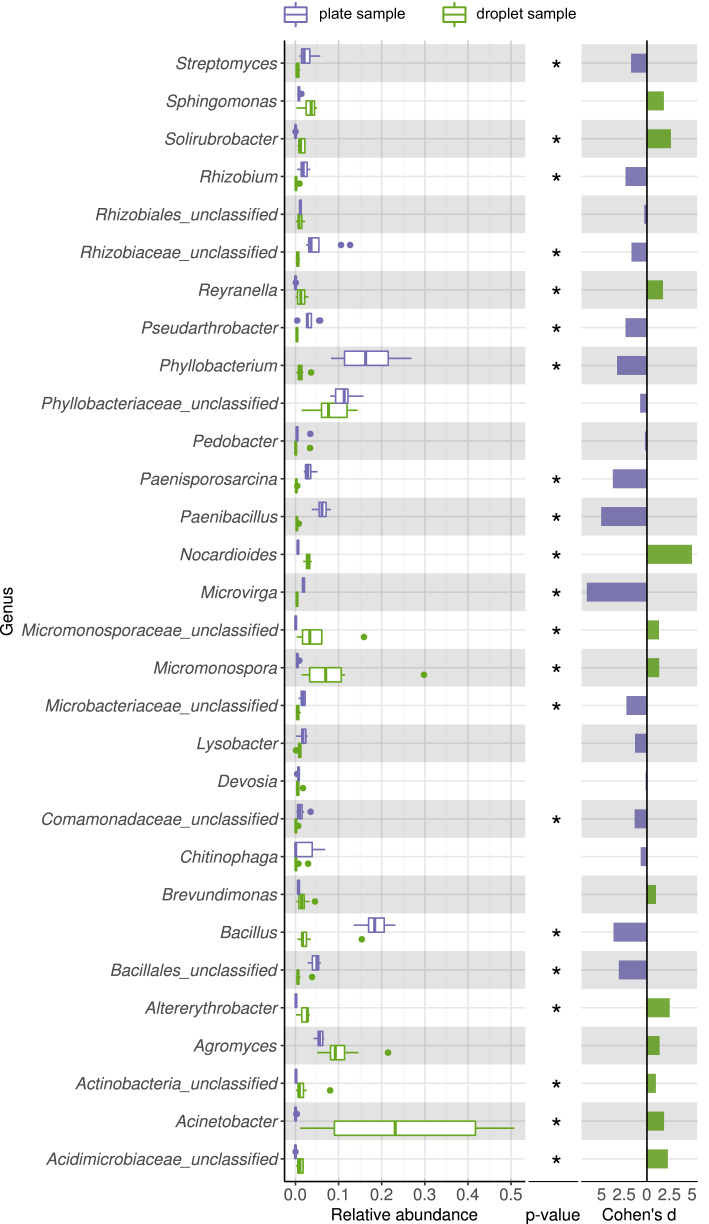
Figure 3. Comparing taxonomic classification on genus rank for replicating cells in agar plate and in-droplet cultivation.
(A) Triangles depict the relative abundances for genera in eight biological replicates per droplet and plate cultivation method. Underlying boxplots show the distribution of data points. Relative abundances of operational taxonomic units (OTU) (evolutionary distance of 0.03) were agglomerated on the genus level (evolutionary distance of 0.1). Displayed are the 10 most abundant genera of in total 238 assigned genera covering 66.8% of all sequences. The 30 most abundant genera covering a total of almost 90% are shown in Figure 3—figure supplement 5. Means of relative abundances were compared by Wilcoxon rank-sum test for each genus (α = 0.05), applying Holm–Bonferroni correction for multiple comparisons. Adjusted p-values are displayed. As effect size, Cohen’s d was computed and plotted as bars to indicate which differences are practically relevant. The direction and the color of the bars depend on the sample type in which the larger mean was found (blue – larger mean in plate samples, green – larger mean in droplet samples). (B) Venn diagram for the taxa on genus level. Of 238 assigned genera, 142 were found in both cultivation methods while 93 were unique to droplet cultivation and three were only observed in plate cultivation. The area of the Venn elements corresponds to the total number of genera found for the cultivation techniques. (C) Venn diagram for the OTU level. Of 1687 OTUs, 677 were found in both cultivation methods, 983 were unique to droplets, and 27 were unique to plates. The area of the Venn elements corresponds to the total number of OTUs found for the cultivation techniques.