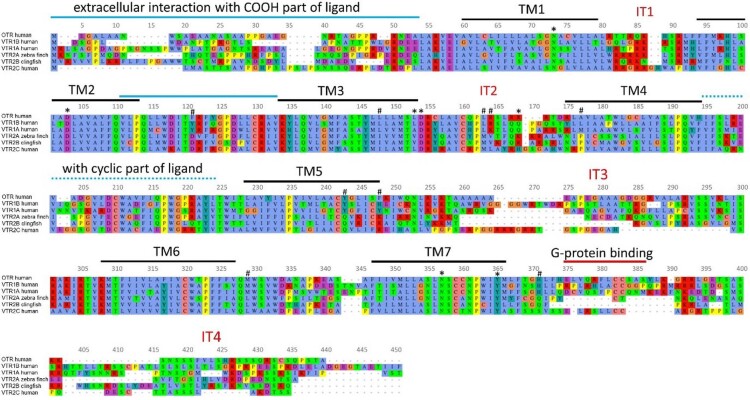
Extended Data Fig. 10. MAFFT alignment of the OTR-VTRs of the best-quality assemblies available (human for OTR, VTR1A, VTR1B and VTR2C; zebra finch for VTR2A; and clingfish for VTR2B).
The MAFFT alignment using the FFT-NS-I parameter was visualized with the MSA viewer46. The identifiers and protein sequences used, along with the alignment file can be found in https://github.com/constantinatheo/otvt. The functional annotation of transmembrane domains (TM) and intracellular loops (IT) and binding domains is based on findings with OTR47. Amino acids marked with an asterisk are the OT polar-interacting sites to the receptor; amino acids marked with a # are differences between the VTR1 and VTR2 subfamilies. Colour coding of the amino acids is according to Clustal X (blue, hydrophobic; red, positive charge; green, polar; pink, cysteines; orange, glycines; yellow, prolines; cyan, aromatic; http://www.jalview.org/help/html/colourSchemes/clustal.html).