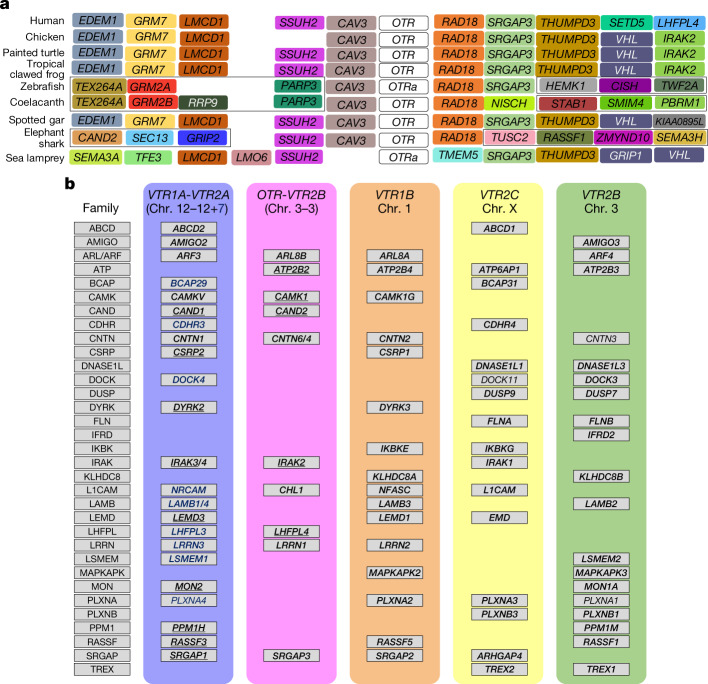
Fig. 2. Interspecies and intraspecies synteny analyses.
a, Example of interspecies ten-gene microsynteny for OTR across vertebrates. Same colour, orthologous genes. Black boxes, genome rearrangements. OTRa in the sea lamprey and zebrafish is orthologous to OTR in all other vertebrates. Human OTR is currently known as OXTR; tropical clawed frog otr is currently known as oxtr. b, Intraspecies 10-Mb macrosynteny among 6 chromosomes (block colours) for all OTR-VTR gene regions in humans whether present (OTR, VTR1A, VTR1B and VTR2C) or deleted (VTR2A and VTR2B). Gene families are listed alphabetically on the left. In the blue column, underlined genes were found within a 10-Mb window of VTR1A on chromosome 12. In the pink column, underlined genes were found within a 10-Mb window of OTR on chromosome 3; genes in black bold were found within a 10-Mb window of the deleted VTR2A on chromosome 12 or (in blue bold) 7, or within the 10-Mb window of the deleted VTR2B on chromosome 3. Orange column, all genes listed (in black bold) were found within a 10-Mb window of VTR1B on chromosome 1 (orange block). Yellow column, all genes listed (in black bold) were found within a 10-Mb window of VTR2C on chromosome X (yellow block). Green column, an alternative syntenic territory of VTR2B (green) was also found at a different location of chromosome 3. Genes not in bold are found outside of the strict 10-Mb window, but are on the same chromosome as the respective OTR-VTR gene.