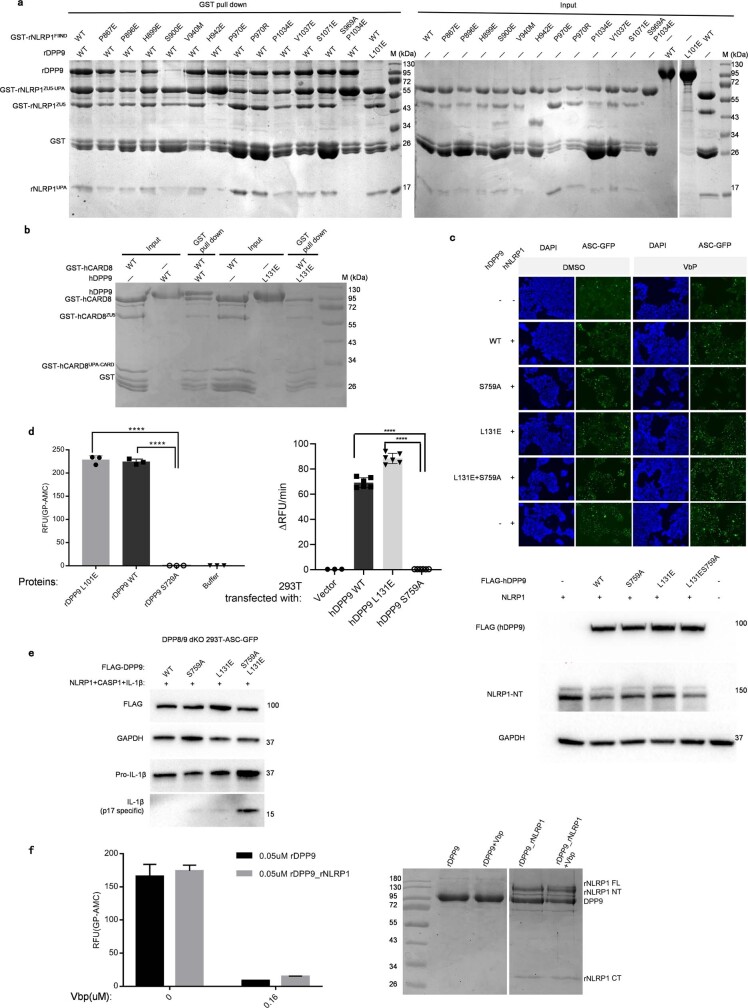
Extended Data Fig. 7. DPP9 interacts with FIIND from rNLRP1 and CARD8.
a, Mutagenesis analysis of the ZU5-binding site of rDPP9. N-terminally GST-fused wild-type or mutant rNLRP1 FIIND and rDPP9 were individually purified from insect cells. Wild-type or mutant rNLRP1 FIIND was used to pull down rDPP9 (including rDPP9(L101E)) with GS4B resin. After extensive washing, proteins bound to the GS4B resin were visualized by SDS–PAGE followed by Coomassie-blue staining. b, The interaction of hDPP9 with CARD8. N-terminally GST-fused CARD8 was used to pull down non-tagged wild-type or mutant hDPP9 in vitro. c, Top, representative images of DPP8/DPP9 double-knockout ASC–GFP 293T cells transfected with hNLRP1 and wild-type hDPP9 or hDPP9 mutants. Bottom, expression of wild-type and DPP9 mutants in DPP8/DPP9 double-knockout 293T cells. The soluble lysate was blotted for DPP9, NLRP1 or GAPDH. d, Mutation of L101 of rDPP9 (left) or its equivalent L131 of hDPP9 (right) has no effect on the protease activity. Each experiment was repeated at least three times. One-way ANOVA, ****P < 0.0001. RFU, relative fluorescence units. e, DPP8/DPP9 double-knockout 293T cells were co-transfected with 3×Flag hDPP9 variant together with CASP1-Myc and pro-IL-1β. ASC–GFP speck formation was monitored for consistency with that in c. Lysates were analysed by immunoblotting. f, Left, inhibition of the protease activity of rDPP9 and rNLRP1–rDPP9 by VbP. Right, SDS–PAGE analyses of the rDPP9 and rNLRP1–rDPP9 complexes used in the activity assays. See Supplementary Figs. 2 and 4 for gel raw data.