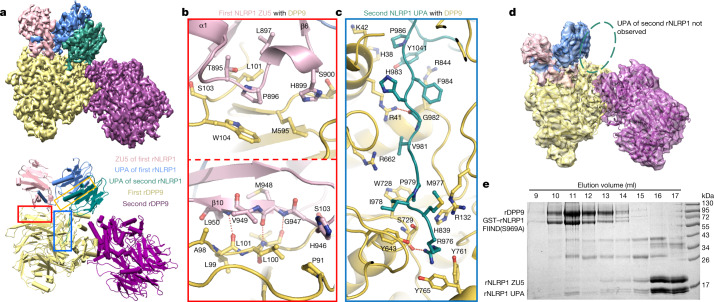
Fig. 2. Assembly mechanism of the 2:1 rNLRP1–rDPP9 complex.
a, Top, the final cryo-EM density of the rNLRP1–rDPP9 complex at 3.18 Å. Colour codes for domain structures are indicated. Bottom, model of the 2:1 rNLRP1–rDPP9 complex. The three interfaces that mediate the rNLRP1–rDPP9 interaction are shown in coloured boxes. Red, ZU5-binding site; blue, UPA-binding site; yellow, UPA–UPA-binding site. b, Detailed interactions between ZU5 and rDPP9 at the ZU5-binding site within the red-framed region in a. Hydrogen-bonding interactions are indicated by red dashed lines. c, Detailed interactions between UPA and rDPP9 at the UPA-binding site within the blue-framed region in a. d, 3D reconstruction of the rNLRP1 FIIND–CARD(S969A)–rDPP9 complex. Highlighted within the open ellipse is the vacant UPA-binding site. e, The ZU5 subdomain is displaced after interaction of the second rNLRP1 with rDPP9. The preformed rNLRP1 FIIND(S969A)–rDPP9 complex was incubated with fully autoprocessed rNLRP1 FIIND and analysed by gel-filtration experiments. Protein fractions from the gel-filtration assay were visualized by SDS–PAGE followed by Coomassie-blue staining. See Supplementary Fig. 1 for gel raw data.