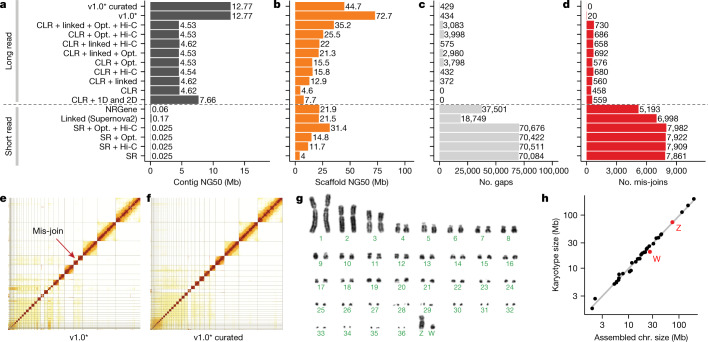
Fig. 1. Comparative analyses of Anna’s hummingbird genome assemblies with various data types.
a, Contig NG50 values of the primary pseudo-haplotype. b, Scaffold NG50 values. c, Number of joins (gaps). d, Number of mis-join errors compared with the curated assembly. The curated assembly has no remaining conflicts with the raw data and thus no known mis-joins. *Same as CLR + linked + Opt. + Hi-C, but with contigs generated with an updated FALCON17 version and earlier Hi-C Salsa version (v2.0 versus v2.2; Supplementary Table 2) for less aggressive contig joining. e, f, Hi-C interaction heat maps before and after manual curation, which identified 34 chromosomes. Grid lines indicate scaffold boundaries. Red arrow, example mis-join that was corrected during curation. g, Karyotype of the identified chromosomes (n = 36 + ZW), consistent with previous findings70. h, Correlation between estimated chromosome sizes (in Mb) based on karyotype images in g and assembled scaffolds in Supplementary Table 4 (bCalAna1) on a log–log scale. v1.0, VGP assembly v1.0 pipeline; linked, 10X Genomics linked reads; Hi-C, Hi-C proximity ligation; 1D, 2D, Oxford Nanopore long reads; NRGene, NRGene paired-end Illumina reads; SR, paired-end Illumina short reads.