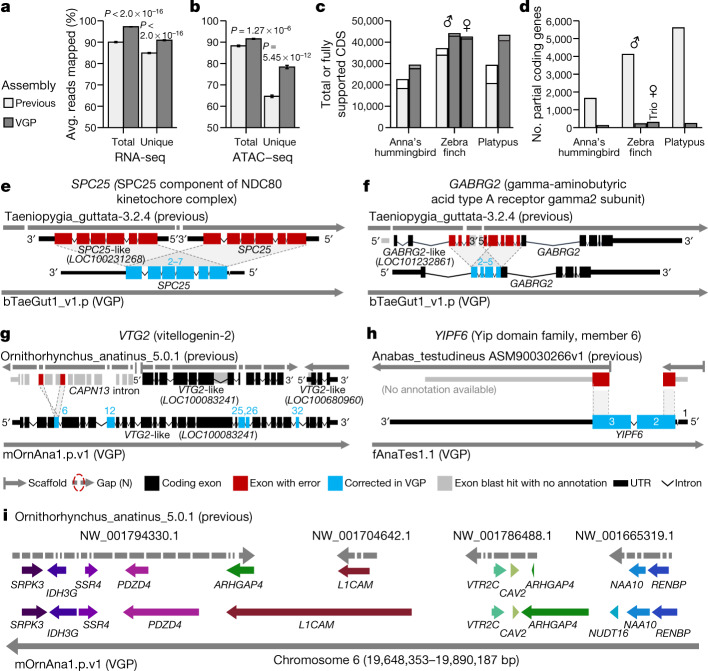
Fig. 3. Improvements to alignments and annotations in VGP assemblies relative to prior references.
a, b, Average percentage of RNA-seq transcriptome samples (a; n = 44, mean ± s.e.m.) and ATAC–seq genome reads (b; n = 12) that align to the previous and VGP zebra finch assemblies. Unique reads mapped to only one location in the assembly. Total is the sum of unique and multi-mapped reads. P values are from paired t-test. c, d, Total number of coding sequence (CDS) transcripts (full bar) and portion fully supported (inner bar) (c) and the number of RefSeq coding genes annotated as partial (d) in the previous and VGP assemblies using the same input data. e–h, Examples of assembly and associated annotation errors in previous reference assemblies corrected in the new VGP assemblies. See main text for descriptions. i, Gene synteny around the VTR2C receptor in the platypus shows completely missing genes (NUDT16), truncated and duplicated ARHGAP4, and many gaps in the earlier Sanger-based assembly compared with the filled in and expanded gene lengths in the new VGP assembly. Assembly accessions are in Supplementary Table 19.