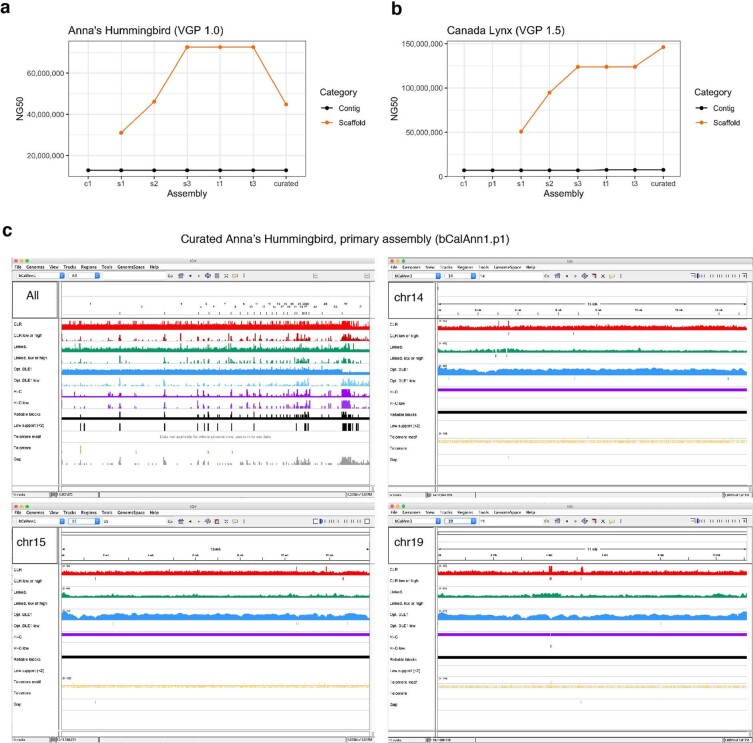
Extended Data Fig. 1. Assessment of completeness of the Anna’s hummingbird assembly.
a, b, Steps and NG50 continuity values of the VGP assembly pipeline that gave the highest quality assembly for Anna’s hummingbird (a) and Canada lynx (b) in this study. The specific steps are outlined further in Extended Data Fig. 2a, and Methods. c, Whole-genome alignment of CLR (red), linked reads (green), optical maps (blue), and Hi-C reads (purple) of the Anna’s hummingbird, along with telomere motif (TTAGGG and its reverse complement, yellow) and gaps (grey) using Asset software103. For each data type, the first row shows the mapped coverage, and the second shows the number of counts of low coverage or signs of collapsed repeats. Larger chromosomal scaffolds (1–19) have fewer gaps and low coverage or collapsed regions compared with the micro chromosomes (20–33). Chromosomes 14, 15 and 19 of the Anna’s hummingbird were the most structurally reliable scaffolds, having only one gap each with no low-support regions. We defined reliable blocks as those supported by at least two technologies. Reliable blocks excluded regions with structural assembly errors, such as collapsed repeats or unresolved segmental duplications. Low-support regions are those where the reliable blocks row has a peak.