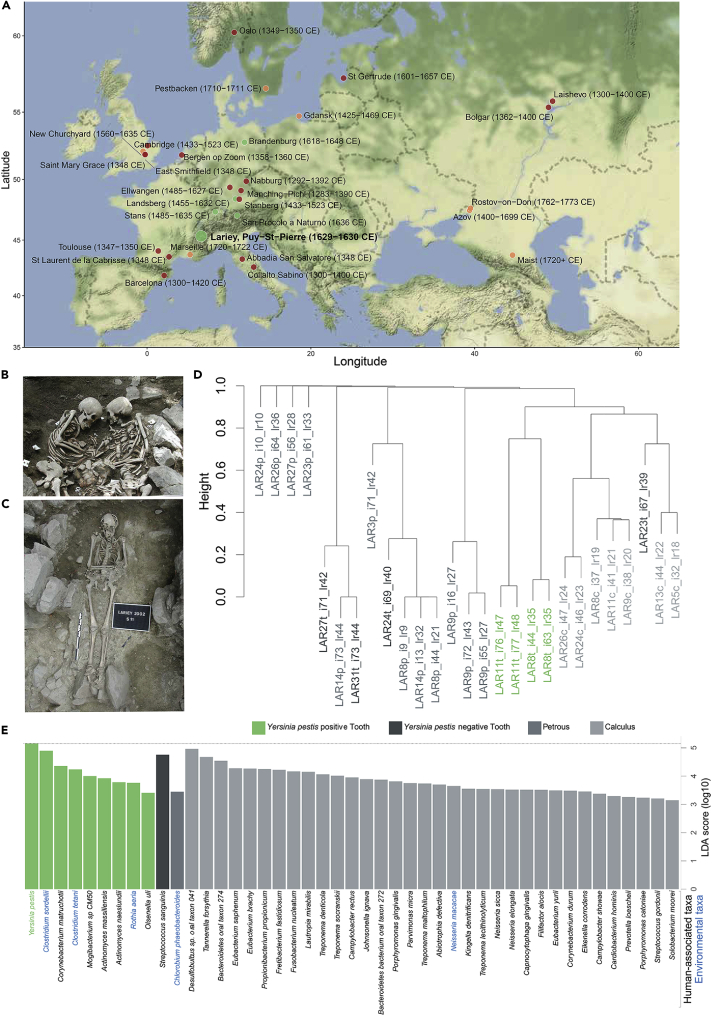
Figure 1.
Sample information and metagenomic analyses
(A) Map showing the location and temporal range of the Lariey-Puy-Saint-Pierre cemetery together with previously published second pandemic plague genomes. Colors are indicated with respect to Spyrou et al. (2019b) and according to the main phylogenetic clusters shown on Figure 3.
(B) Multiple burial showing the LAR8 individual (right).
(C) Simple burial showing the LAR11 individual.
(D) Hierarchical clustering dendrogram of Bray-Curtis distances between MetaPhlAn2 (Truong et al., 2015) bacterial abundance profiles (10,000 bootstrap pseudo-replicates) and disregarding abundances below 1%. All clusters are supported with a pvclust (Suzuki and Shimodaira, 2006) approximately unbiased p value of 100.
(E) LEfSe (Segata et al., 2011) Linear Discriminant Analysis indicating those microbial species with most contrasted abundance patterns (LDA scores >3). Higher log10-LDA scores identify those bacterial species contributing the most to the differences in the metagenomic content of teeth positive or negative for Yersinia pestis, petrosal and dental calculus remains. The source of each species was predicted on the basis of the literature and conservatively categorized as ‘environmental’ whenever both sources were likely.
See also Figures S1 and S2.