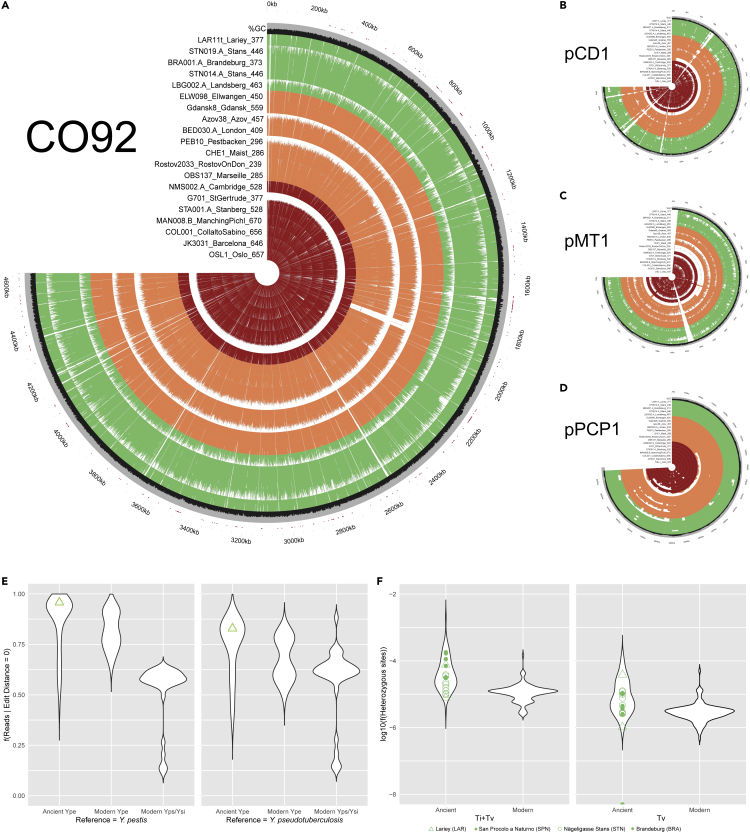
Figure 2.
Plague chromosome and plasmid sequence coverage
(A) Coverage and %GC variation along 1,000 bp windows along the CO92 plague reference genome ( GenBank: NC_003143.1) (Parkhill et al., 2001). Average depth-of-coverage was calculated using Paleomix (Schubert et al., 2014). %GC composition was calculated using seqtk (https://github.com/lh3/seqtk). Circular plots were traced using the circlize library in R (Gu et al., 2014) (max = 10-fold). In case multiple genomes are available for one site, the genome showing maximal coverage is shown.
(B) Coverage and %GC variation along 100 bp windows along the pCD1 plague plasmid (GenBank: NC_003131.1).
(C) Same as panel B, except that the pMT1 plasmid is shown (GenBank: NC_003134.1).
(D) Same as panel B, except that the pPCP1 plasmid is shown (GenBank: AL109969.1).
(E) Edit distance profiles obtained when mapping the sequence data underlying the LAR8 and LAR11 genomes and a comparative panel of 78 ancient and 155 modern Yersiniapestis (Ype), Yersinia pseudotuberculosis (Yps) and Yersinia similis (Ysi) sequence datasets aligned against the Ype and Yps reference genomes, respectively (Table S2). Edit distance distributions were considered as long as based on a minimum of 500 reads.
(F) Heterozygosity profiles of the LAR8 and LAR11 genomes and a comparative panel of 78 ancient and 129 modern Yersinia pestis sequence datasets aligned against the CO92 Yersinia pestis reference chromosome (Table S2). Transition SNPs were disregarded to account for the differential rates of postmortem DNA damage amongst the comparative panel.