Figure 5.
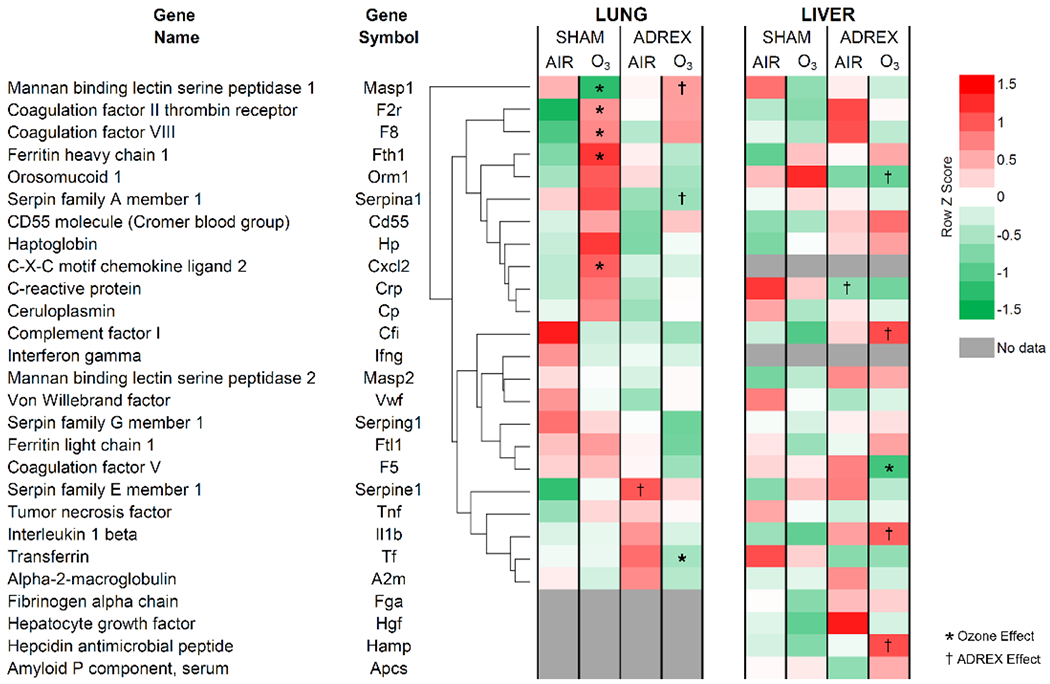
Altered expression of APP genes in the lung and liver after 1 day of O3 exposure in SHAM and ADREX rats (Study 2). Selected APR genes were identified from RNAseq Illumina data set for lung and liver from air or O3 (1 ppm for lung and 0.8 ppm for liver; each for 4 hr) exposed SHAM and ADREX rats (n=4-5/group). Normalized gene expression values for selected APP genes from each tissue were separated from RNAseq data set, and heat maps of Row z scores were constructed. For lung tissue, average linkage clustering and Euclidean distance measurement was employed, and then liver genes were arranged in the same sequence as lung without clustering for ease of comparison. Red and green indicate increased and decreased gene expression, respectively. Significant changes in expression due to O3 exposure is indicated by an *, and due to surgical treatment is indicated by a † (adjusted-p-value-cut-off of 0.05).
