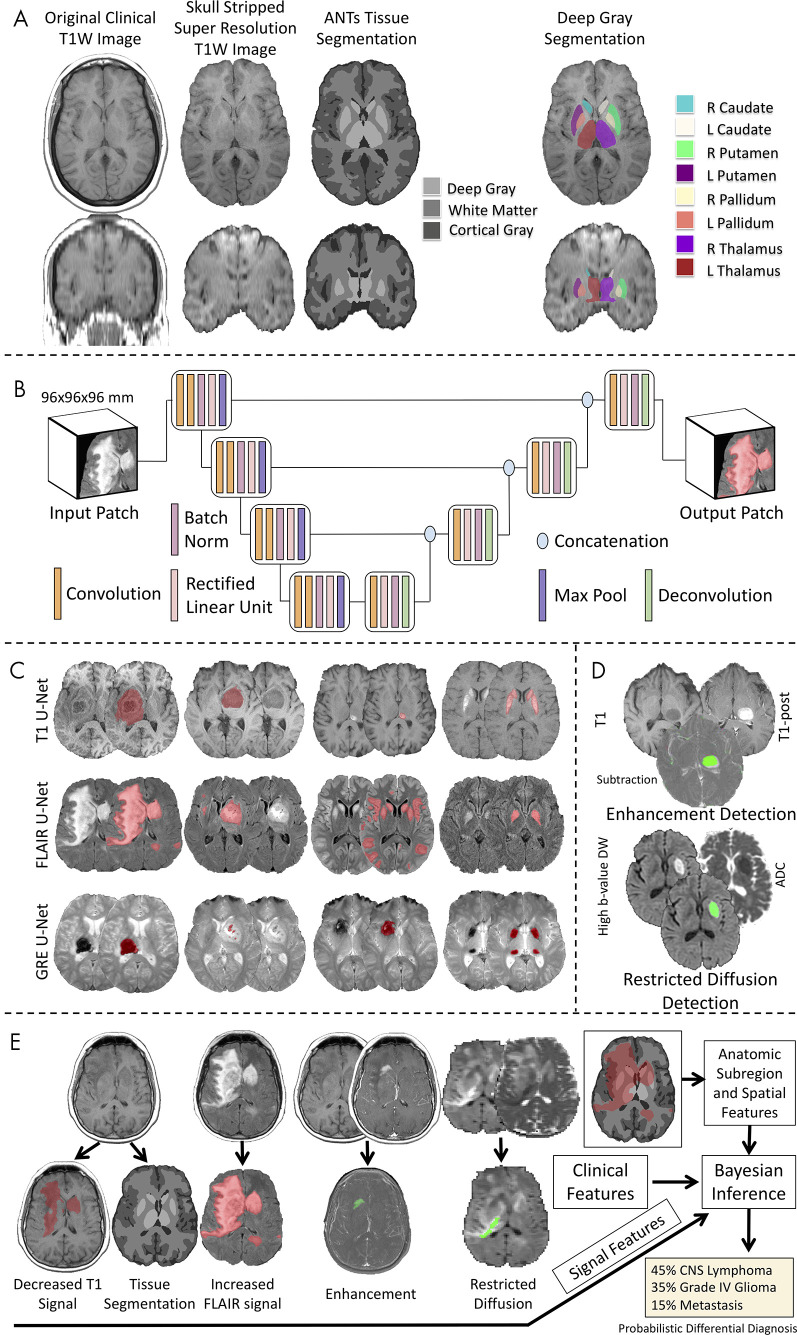
Figure 3:
Workflow of the image processing pipeline. A, Atlas-based neuroimaging processing pipeline for tissue segmentation and deep gray matter parcellation. T1-weighted (T1W) axial (upper row) and coronal (lower row) MRI scans were up-sampled and skull-stripped (second column) before tissue segmentation with the Advanced Normalization Tools (ANTs) pipeline (third column) and parcellation of deep gray matter structures (fourth column). B, Diagrammatic overview of the custom three-dimensional U-Net architecture for abnormal signal detection. C, Examples of U-Net–based segmentations for T1-weighted (T1, first row), T2-weighted fluid-attenuated inversion recovery (FLAIR, second row), and gradient-recalled echo (GRE, third row) MRI scans of test case. D, Example of T1-weighted (T1), T1-weighted postcontrast (T1-post), and a subtraction of the T1-weighted image from the T1-weighted postcontrast image with detected areas of abnormal enhancement (green) and high b value diffusion-weighted (DW) and apparent diffusion coefficient (ADC) images with detected areas of restricted diffusion (green). E, Example of correctly diagnosed central nervous system (CNS) lymphoma processed through the full pipeline with signal, anatomic subregion, and spatial features (derived from abnormal signal segmentations overlaid on tissue segmentation maps) combined with clinical features into a Bayesian inference system to derive a probabilistic differential diagnosis.