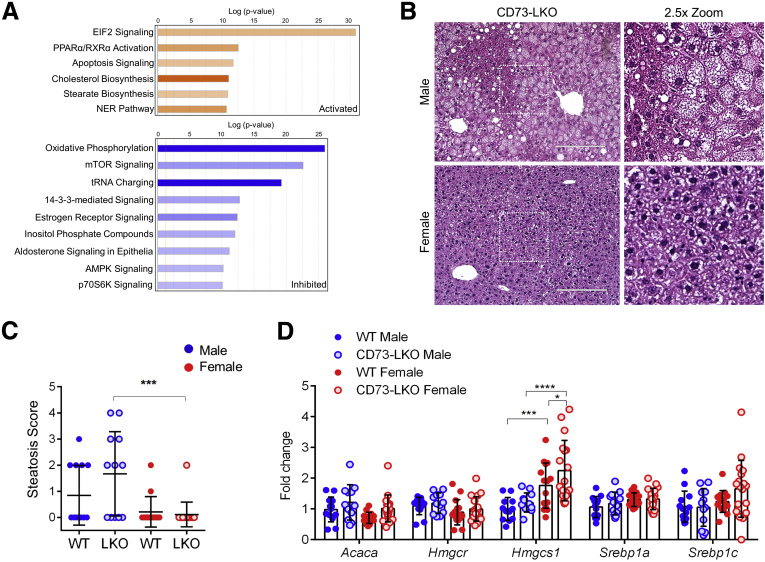
Figure 5.
Metabolic imbalance and increased steatosis in male CD73-LKO mouse hepatocytes and livers. (A) Ingenuity pathway analysis of proteomic results showing activated (orange) and inhibited (blue) pathways in freshly isolated hepatocytes from male CD73-LKO mice relative to WT controls. (B) H&E staining of formalin-fixed liver sections. Magnified view of boxed areas in the left panel comparing the presence of steatosis in male and female CD73-LKO mice. Scale bar: 200 μm. (C) Quantification of blinded histologic scoring of microvesicular and macrovesicular steatosis. Scoring: 0, none; 1, mild; 2, moderate; 3, severe (scores were averaged for animals with microsteatosis and macrosteatosis). Males: n = 13 WT; n = 12 CD73-LKO; females: n = 14 WT; n = 18 CD73-LKO. ∗∗∗P < .001, 1-way analysis of variance. (D) Gene expression analysis of lipid metabolism genes Acaca, Hmgcr, Hmgcs1, Srebp1a, and Srebp1c using total liver messenger RNA in male and female WT and CD73-LKO mice. n = 13 WT male, n = 12 LKO male, n = 14 WT female, n = 18 LKO female. ∗P < .05, ∗∗∗P < .001, ∗∗∗∗P < .0001, 2-way analysis of variance. Error bars represent SD. EIF2, eukaryotic initiation factor 2; NER, nucleotide excision repair; PPARα/RXRα, peroxisome proliferator-activated receptor α/retinoid X receptor α; tRNA, transfer RNA.