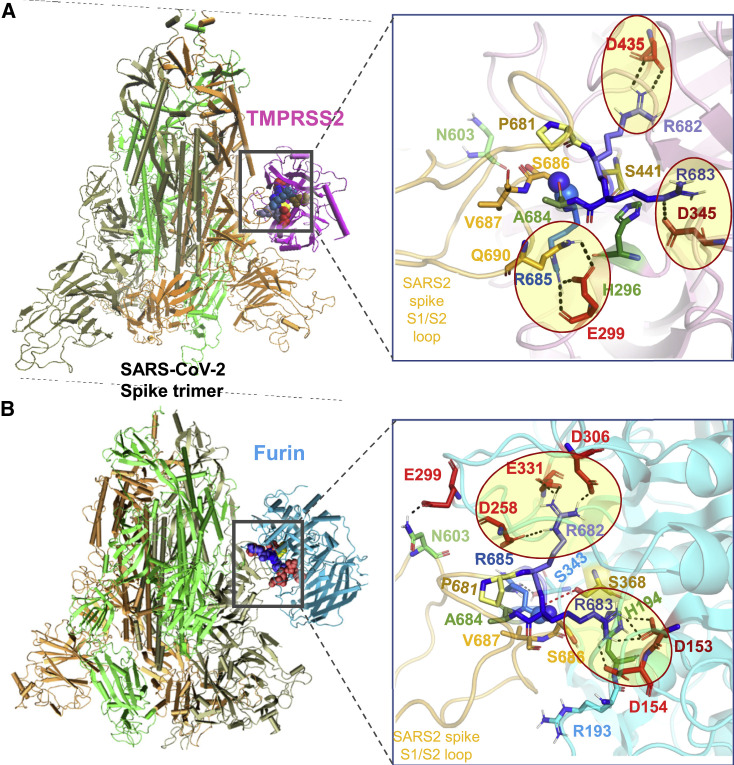
Figure 5.
Binding poses of human proteases TMPRSS2 and furin to SARS-CoV-2 S protein
(A and B) Structural models for the SARS-CoV-2 S protein complexed with (A) TMPRSS2 and (B) furin, obtained from docking simulations followed by refinements. An overview (left) and a zoomed-in view (right) are shown in each case. The arginines in the S1/S2 loop P681RRARS686 are shown in different shades of blue, and their interaction partners (acidic residues) in the proteases are shown in red. Spheres (right) highlight the R685↑S686 peptide bond. The TMPRSS2 catalytic triad residues are S441 (yellow), H296 (green), and D345 (dark red). Their counterparts in furin are S368, H194, and D153. Note the short distance between the carbonyl carbon of R685 and the hydroxyl oxygen of S441 of TMPRSS2 (3.5 Å) or S368 of furin (3.1 Å). Black dashed lines show interfacial polar contacts and salt bridges, and those including the S1/S2 loop arginines are highlighted by ellipses.