Abstract
Background
Duchenne muscular dystrophy (DMD) leads to progressive cardiomyopathy. Detection of myocardial fibrosis with late gadolinium enhancement (LGE) by cardiovascular magnetic resonance (CMR) is critical for clinical management. Due to concerns of brain deposition of gadolinium, non-contrast methods for detecting and monitoring myocardial fibrosis would be beneficial.
Objectives
We hypothesized that native T1 mapping and/or circumferential (εcc) and longitudinal (εls) strain can detect myocardial fibrosis.
Methods
156 CMRs with gadolinium were performed in 66 DMD boys and included: (1) left ventricular ejection fraction (LVEF), (2) LGE, (3) native T1 mapping and myocardial tagging (εcc-tag measured using harmonic phase analysis). LGE was graded as: (1) presence/absence by segment, slice, and globally; (2) global severity from 0 (no LGE) to 4 (severe); (3) percent LGE using full width half maximum (FWHM). εls and εcc measured using feature tracking. Regression models to predict LGE included native T1 and either εcc-tag or εls and εcc measured at each segment, slice, and globally.
Results
Mean age and LVEF at first CMR were 14 years and 54%, respectively. Global εls and εcc strongly predicted presence or absence of LGE (OR 2.6 [1.1, 6.0], p = 0.029, and OR 2.3 [1.0, 5.1], p = 0.049, respectively) while global native T1 did not. Global εcc, εls, and native T1 predicted global severity score (OR 2.6 [1.4, 4.8], p = 0.002, OR 2.6 [1.4, 6.0], p = 0.002, and OR 1.8 [1.1, 3.1], p = 0.025, respectively). εls correlated with change in LGE by severity score (n = 33, 3.8 [1.0, 14.2], p = 0.048) and εcc-tag correlated with change in percent LGE by FWHM (n = 34, OR 0.2 [0.1, 0.9], p = 0.01).
Conclusions
Pre-contrast sequences predict presence and severity of LGE, with εls and εcc being more predictive in most models, but there was not an observable advantage over using LVEF as a predictor. Change in LGE was predicted by εls (global severity score) and εcc-tag (FWHM). While statistically significant, our results suggest these sequences are currently not a replacement for LGE and may only have utility in a very limited subset of DMD patients.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12968-021-00736-1.
Keywords: Duchene muscular dystrophy, Late gadolinium enhancement, Circumferential strain, Native T1, Cardiac fibrosis
Introduction
Duchenne muscular dystrophy (DMD) leads to progressive skeletal and cardiac myopathy and affects up to 1 in 4700 live male births [1]. Disease progression is variable, but all boys will have cardiovascular manifestations by 18 years of age [2]. There is no cure for DMD. While cardiovascular therapeutic options are limited, early therapy with angiotensin converting enzyme inhibitors and/or aldosterone inhibitors has demonstrated delayed mortality [3, 4]. Some boys with DMD have earlier onset of cardiovascular disease or more rapid progression, requiring either earlier initiation or more rapid intensification of therapy, resulting in early heart failure and death.
Cardiovascular imaging is necessary to detect both early onset and rapid progression of cardiomyopathy. While transthoracic echocardiography has been the mainstay of cardiovascular evaluation in DMD, the field has increasingly shifted to cardiovascular magnetic resonance (CMR) [5]. This is partially because CMR provides superior image quality, a significant advantage in a patient population with challenging acoustic windows [6]. CMR also allows for assessment of fibrosis with late gadolinium enhancement (LGE), which can precede left ventricular (LV) dysfunction and which predicts morbidity and mortality in multiple disease processes, including DMD [7–11]. Clinical decision making is often based on LGE in boys with DMD. Tandon and colleagues have shown that LV ejection fraction (LVEF) declines once LGE is present but not before [11]. Thus, LGE can be an early sign of impending LV dysfunction. In addition, given the increased mortality associated with LGE, a more rapid progression of LGE in the setting of stable or mild changes in LVEF is treated with more aggressive medical therapy. The benefit of LGE is the information it provides in addition to LVEF, especially in a patient with stable ventricular function. However, LGE requires administration of gadolinium contrast agents. While these agents have an excellent safety profile in patients without renal disease, recent reports suggest that renal disease may be under-recognized in older DMD patients [12], coupled with growing concern over possible long-term brain deposition after repeated scans has raised concerns about long-term safety in children and young adults [8, 13].
Recently, mapping of the longitudinal time constant, T1, has gained popularity for myocardial tissue characterization. Native T1 maps can be performed without contrast administration, while extracellular volume (ECV) maps require a combination of native T1 and post-contrast T1 maps as well as a recent hematocrit [14–16]. Native T1 times are elevated in the presence of myocardial fibrosis, and correlates with LGE in other disease processes [17–20]. We previously demonstrated that native T1 is elevated in DMD patients compared with healthy subjects [21]. Myocardial strain has also been used to determine the presence or absence of LGE [22]. We hypothesized that native T1 and/or circumferential (εcc) and longitudinal strain (εls) can be used as surrogates for LGE in DMD subjects, allowing for less frequent gadolinium contrast administration.
Methods
Patient selection
Subjects were drawn from prospective observational studies, all of which were approved by the Vanderbilt Institutional Review Board. All patients signed approved consents or assents. Inclusion criteria were the following: (1) DMD diagnosed phenotypically and confirmed with either genetic testing or muscle biopsy, (2) at least one CMR performed. Exclusion criteria were: (1) other genetic diagnosis in addition to DMD, (2) renal disease or other diagnosis precluding CMR with contrast, (3) no LGE imaging performed on CMR, (4) inadequate T1 map quality in all three imaging slices on all available CMRs. As the incidence of renal disease is low in the general pediatric and DMD populations, creatinine is not usually assessed unless there is a clinical concern. However, screening blood chemistries including creatinine are performed clinically, particularly for patients on angiotensin converting enzyme inhibitors or aldosterone inhibitors. Of the patients enrolled in our studies, there were a total of 8 CMRs where a patient did not receive contrast and so were removed from our analysis. Of those, one was because of anxiety in the scanner so the patient was removed early, 7 were in patients in whom no intravenous access could be obtained or the intravenous catheter infiltrated.
CMR protocol
CMR was performed on a 1.5T CMR system (Avanto, Siemens Healthineers, Erlangen, Germany). Functional imaging was performed as previously described using balanced steady state free-precession (bSSFP) images in a short axis stack [23]. Intravenous Gd-DTPA contrast (gadopentate dimeglumine, Magnevist®, Bayer Healthcare Berlin, Germany or gadobutrol, Gadovist®, Bayer Healthcare) was administered through a peripheral intravenous line at a dose of 0.2 mmol/kg. LGE imaging was performed using: (1) single shot (bSSFP) and segmented (turboFLASH) inversion recovery with optimized inversion recovery to null the signal from the myocardium, as well as phase sensitive inversion recovery (PSIR) bSSFP with an inversion time of 300 ms.
Myocardial tagging was performed in the LV short axis at the base, papillary muscles, and apex using a segmented k-space fast gradient echo sequence with electrocardiographic (ECG) triggering. Grid tagging was performed with a spacing of 8 mm and 9–13 phases. Typical imaging parameters included: slice thickness 6–8 mm, field-of-view 340 mm × 340 mm, matrix size 256 × 192, and minimum echo time and repetition time. The sequences were breath-holds and parallel imaging with generalized autocalibrating partially parallel acquisition (GRAPPA) with an acceleration factor of two. Breath-held modified Look-Locker inversion recovery (MOLLI) sequences (investigative) were performed prior to contrast administration in the LV short axis at the base, mid-LV, and apex at the same slice locations as the tagging [24]. MOLLI sequences were motion-corrected, ECG-triggered images obtained in diastole with typical imaging parameters: non-selective inversion with a 35° flip angle, single shot bSSFP imaging, initial inversion time of 120 ms with 80 ms increments, field-of-view 340 × 272 mm2, matrix size 256 × 144, slice thickness 8 mm, voxel size 1.3 × 1.9 × 8.0 mm3, TR/TE 2.6 ms/1.1 ms, parallel imaging factor of 2. The matrix size was decreased to 192 × 128 for heart rates > 90 (approximate voxel size 1.8 × 2.1 × 8 mm3; 72% of studies were performed with this smaller matrix size). The pre-contrast MOLLI acquired 5 images after the first inversion with the equivalent of a 3 s pause followed by 3 images after the second inversion, or 5(3s)3 (the number of heartbeats used for recovery was varied depending on the average heart rate just prior to T1 mapping: 3 beats for heart rate of 60, 4 beats for heart rate of 80, 5 beats for heart rate of 100, and 6 beats for heart rate of 120) [25]. Motion correction as described by Xue, et al. was performed and a T1 map was generated on the scanner [26]. A goodness of fit map was also performed at the time of the scan to evaluate data quality.
CMR post-processing
LV volume, mass, and function were calculated as previously described [27]. The presence or absence of LGE was qualitatively assessed by one reader (JS). Global severity score was calculated as described previously, with a range of 0 (no LGE) to 4 (severe LGE) [10, 28]. A second reader (FR) performed a blinded analysis of 30 CMRs to evaluate reproducibility of global severity score. Percent of scar was calculated with the full width half maximum (FWHM) method using QMass (Medis Medical Imaging Systems, Leiden, The Netherlands) on the PSIR images. Analysis of myocardial tagged images was performed as previously described using harmonic phase (HARP) methodology (Myocardial Solutions, Morrisville, North Carolina, USA) [29]. In brief, a contour or mesh was drawn over the tagged image at peak systole by outlining the epicardium and endocardium. The superior right ventricular (RV) insertion was identified manually. The contours were performed by the same reader (KGD) with verification of each contour by a second reader (JHS) with more than 7 years of experience using the software. The software then automatically calculated the εcc-tag values for each segment (16 segment model) and slice (base, mid-LV, and apex). Our prior work has demonstrated excellent reproducibility for harmonic phase magnetic resonance (HARP) analysis [6].
Qstrain (Medis Medical Imaging) was used to calculate feature tracking circumferential and longitudinal strain (εcc and εls, respectively). For εcc, cine images in the LV short axis at the base, mid-LV, and apex were chosen and for εls, cine images in the long axis 4-chamber, 3-chamber, and 2-chamber views were chosen. Endocardial borders were traced on each image at end diastole and end systole. Images were inspected closely to confirm adequate tracking and segments that did not track were removed from analysis; more than one segment in each slice with poor tracking resulted in the removal of that CMR from analysis. Global and segmental circumferential and longitudinal strain were calculated automatically by the software.
Using Qmaps (Medis Medical Imaging), epicardial and endocardial borders were manually drawn on native T1 maps within the LV mesocardium. The superior RV insertion was identified and automatically divided into segments using the American Heart Association standard 16 segment model [30]. Regions of interest were carefully traced to avoid partial volume averaging with blood-pool or epicardial fat. Based on the T1 mapping consensus statement, areas of LGE were not excluded as these areas were felt to be the most focal areas in a continuum of diffuse extracellular matrix expansion [31].
In patients with adequate maps at the base and mid-LV slices, global myocardial T1 was calculated. Apical slices were not used to minimize errors from partial volume averaging. Imaging artifact was not contoured. For native T1 maps, segments were not included in the analysis if the bounds of the myocardium could not be distinguished from surrounding tissue and blood pool or if image registration was inadequate in those segments. The total number of excluded segments was 112 out of 2018, with 27/750 from the base, 23/864 from the mid-LV, and 62/404 from the apex. The mean myocardial wall thickness of the baseline CMRs was 6.7 ± 1.3 mm at the septum and 6.0 ± 0.9 mm at the free wall. Figure 1 demonstrates representative LGE, native T1, tagged images, and Fig. 2 shows an example of feature tracking analysis.
Fig. 1.
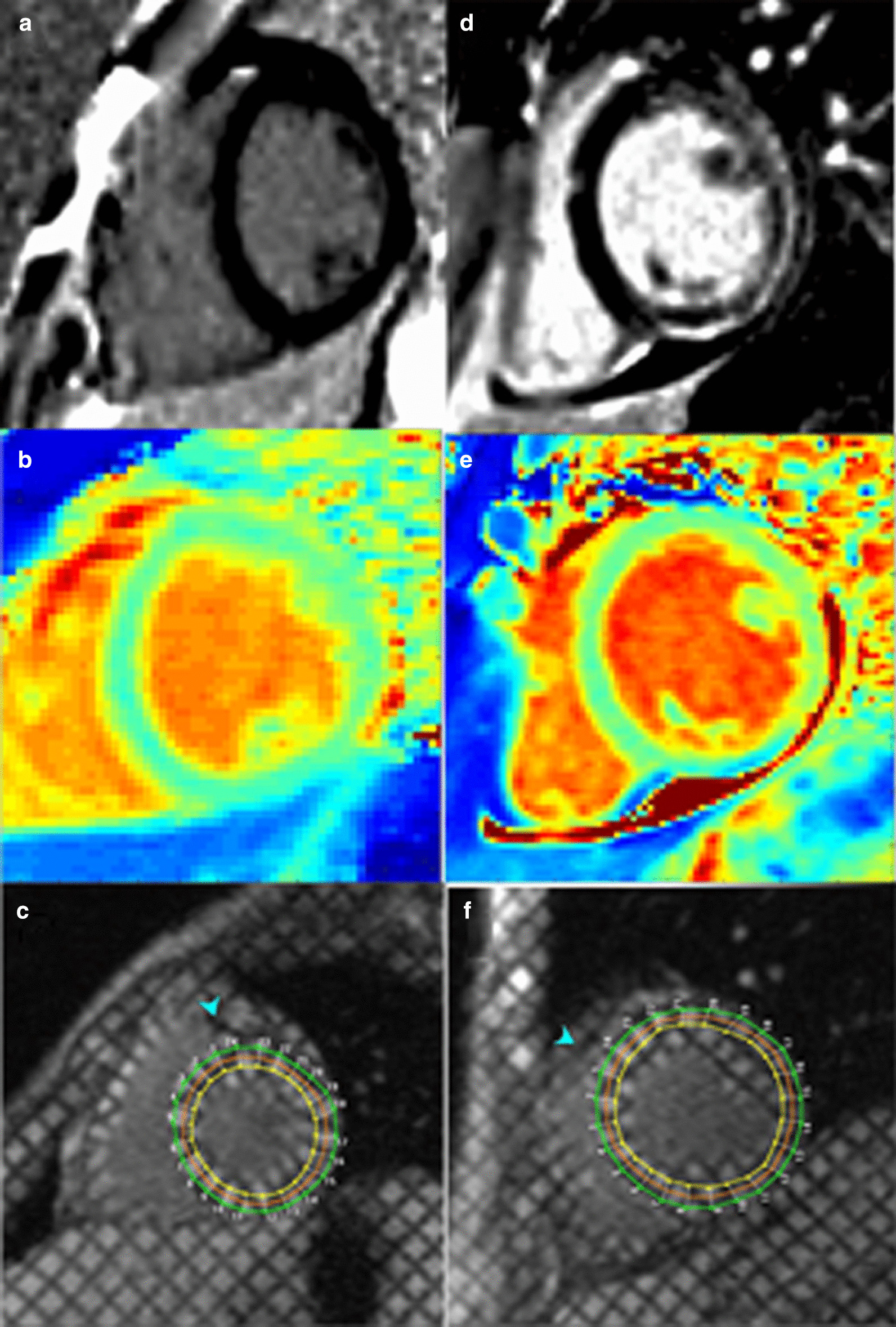
Representative image analysis for T1 and tagging. Representative late gadolinium enhancement (LGE) images (a, d), pre-contrast native T1 maps (b, e), and myocardial tagging (c, f) for patients without (a–c) and with (d–f) LGE
Fig. 2.
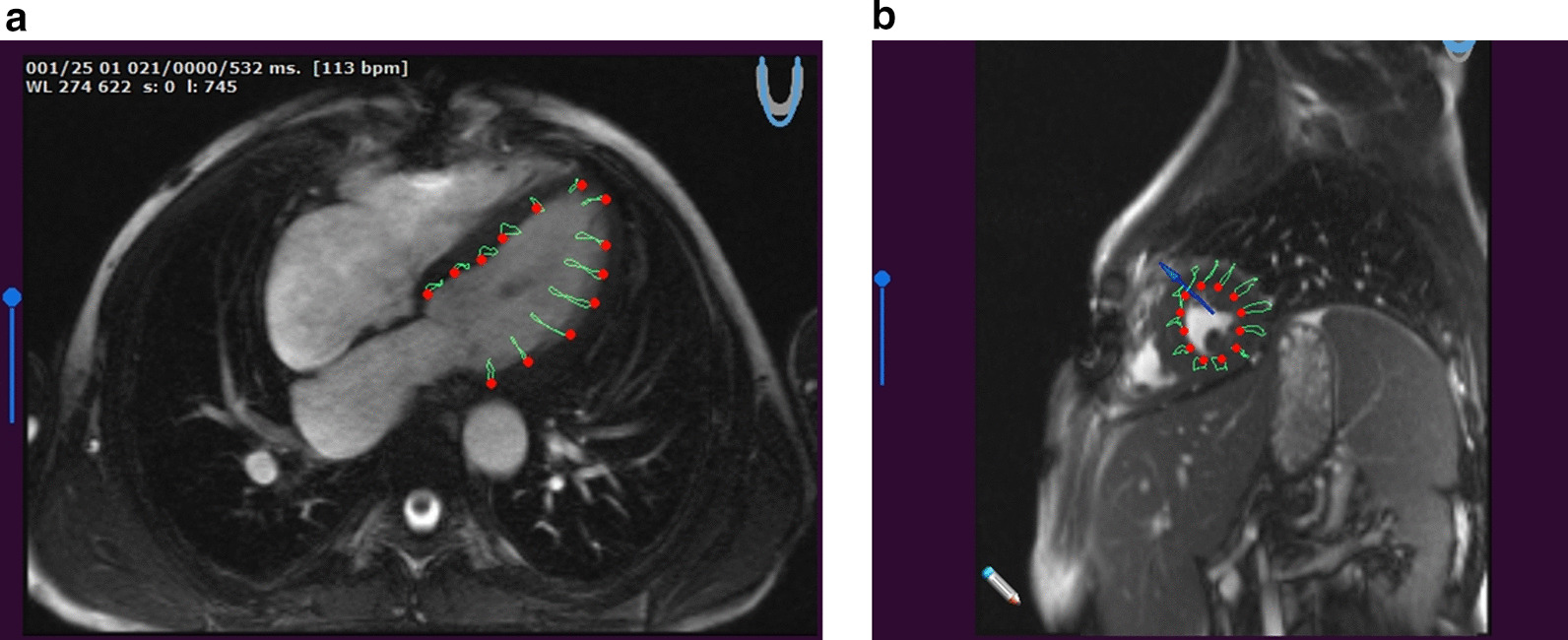
Representative image analysis for feature tracking. Examples of four-chamber view with tracking (a) and short axis for circumferential strain calculation (b)
Statistical analysis
Continuous data are presented as a mean and standard deviation. A missing data summary analysis was performed (Additional file 1: Figure S1). Reproducibility of global severity score was assessed using intraclass correlation coefficient and a weighted kappa. All-cause mortality at 5 years from first CMR in patients with and without LGE was evaluated using Cox regression analysis. Logistic, ordinal logistic, and linear regression models were used for binary outcomes, non-normally distributed continuous outcomes and change of the continuous outcomes respectively using Huber-White sandwich estimator to account for repeated measurements with εls, εcc, and T1 mapping pre-specified as predictors and adjusting for age (and baseline measurements in the change model) as covariates.
Logistic, ordinal, and linear regression models were used to examine longitudinal progression. Progression was defined as: newly developed presence of LGE (logistic regression); any worsening of LGE global severity score (ordinal regression); any worsening of FWHM (linear regression). It should be noted that inclusion of LVEF in these models was considered; however, the decision was made to exclude LVEF and indexed LV end-systolic volume (LVESV) due to significant multicollinearity with strain parameters. We have included examples of univariate models for LVEF and indexed LVESV in Additional file 2: Figure S2). Analyses were performed with R version 3.5.2 (R Foundation for Statistical Computing, Vienna, Austria). Study data were collected and managed using REDCap (Research Electronic Data Capture) electronic data capture tools hosted at Vanderbilt [32].
Results
Demographics and outcomes
A total of 156 CMRs were analyzed from 66 enrolled patients (mean age and LVEF at first CMR were 14 years and 54.3%, respectively). The average total scan time was 40 min. At the time of the CMR, there were 33 patients on corticosteroids and 45 patients on cardiac medications. Seventeen patients were ambulatory. Forty-four patients (70%) were positive for LGE, with 12 patients having global severity score (GSS) = 1, 12 with GSS = 2, 13 with GSS = 3, and 7 with GSS = 4 (Table 1). Reproducibility analysis for GSS demonstrated an intraclass correlation coefficient of 0.99 (p < 0.001) and a weighted kappa of 0.97 (p < 0.001). Patients with LGE had significantly higher all-cause mortality (Fig. 3).
Table 1.
Demographics at the time of their first cardiovascular magnetic resonance scan
| Mean ± SD or N (%) | |
|---|---|
| Age (years) | 14 ± 5 |
| Height (m) | 1.46 ± 0.18 (n = 65) |
| Weight (kg) | 50.1 ± 17.8 |
| BSA (m2) | 1.41 ± 0.32 |
| Ambulatory | 17(26%) |
| Medications at baseline CMR | |
| Corticosteroid | 33 (50%) |
| Angiotensin converting enzyme inhibitor | 33 (50%) |
| Beta blocker | 24 (36%) |
| Aldosterone inhibitor | 4 (6%) |
| Angiotensin receptor blocker | 6(9%) |
| LVEF (%) | 54.3 ± 9.7 |
|
LVEDV (mL) LVEDVI (mL/m2) |
93 ± 42 67 ± 20 (n = 65) |
|
LVESV (mL) LVESVI (mL/m2) |
45 ± 33 32 ± 17 (n = 65) |
|
LV mass (g) Indexed LV mass (g/m2) |
71 ± 27 49 ± 13 (n = 65) |
| Global native T1 (ms) | 1048 ± 44 (n = 54) |
| Global εcc (%) | − 14.1 ± 3.5 (n = 64) |
| Presence of late gadolinium enhancement (LGE) | 44 (70%) |
| Full width half maximum quantification of LGE (%) | 21.3 ± 18.4 (n = 60) |
| GSS = 0 | 19 (30%) |
| GSS = 1 | 12 (19%) |
| GSS = 2 | 12 (19%) |
| GSS = 3 | 13 (21%) |
| GSS = 4 | 7 (11%) |
N = 66 unless otherwise indicated
BSA body surface area, GSS global severity score, LGE late gadolinium enhancement, LV left ventricular, LVEDV left ventricular end-diastolic volume, LVEDVI left ventricular end-diastolic volume index, LVEF left ventricular ejection fraction, LVESV left ventricular end-systolic volume, LVESVI left ventricular end-systolic volume index
Fig. 3.
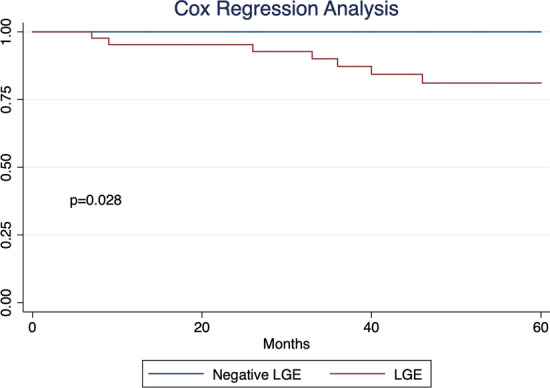
LGE Effect on Duchenne’s muscular dystrophy (DMD) survival. Cox regression survival analysis for our DMD cohort showed significant difference in all-cause mortality in patients with and without LGE
Native T1 and strain predict the presence/absence and extent of LGE at a single time point
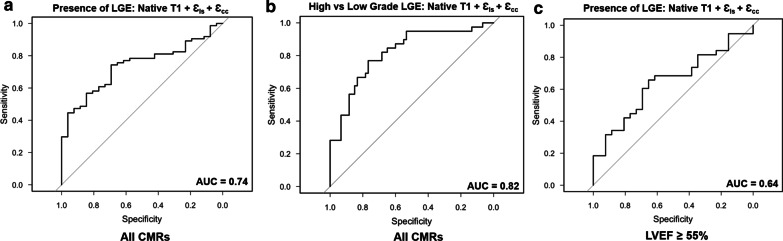
A model including global εls, εcc, and native T1 predicted the presence or absence of LGE, though native T1 failed to reach significance (Table 2), with a correlation statistic (C) = 0.74 and r2 = 0.21, n = 95 (Fig. 4a). εcc, εls, and native T1 all correlated with global severity score (Table 2), however only global εls correlated with percent LGE by FWHM (Table 2). There was not sufficient power to estimate specific cutoffs for the individual variables so combined prediction models for predicting presence/absence of LGE and high grade (GSS ≥ 3) vs low grade (GSS ≤ 2) were created (Fig. 4a, b). Thresholds were determined for the best sensitivity and specificity (with a minimum of 40% in the other) as well as for the optimal sensitivity/specificity combination (Table 3). Sensitivity, specificity, and thresholds for univariate LVEF analysis is included as a reference.
Table 2.
Models for prediction of presence and severity of late gadolinium enhancement (LGE)
| Slice | Factor | Odds ratio and 95% CI | p value | |
|---|---|---|---|---|
| Presence/absence of LGE | ||||
| Base (n = 105) | Native T1 | 2.1 [1.2, 3.8] | 0.012 | |
| εcc | 2.6 [1.1, 6.1] | 0.034 | ||
| εls | 2.3 [1.1, 4.8] | 0.032 | ||
| Mid (n = 133) | Native T1 | 1.2 [0.8, 1.7] | 0.42 | |
| εcc | 2.4 [1.2, 4.7] | 0.009 | ||
| εls | 2.3 [1.4, 3.7] | 0.001 | ||
| Apex (n = 106) | Native T1 | 1.4 [0.7, 2.8] | 0.30 | |
| εcc | 1.9 [0.9, 4.1] | 0.12 | ||
| εls | 1.0 [0.4, 2.6] | 0.93 | ||
| Global (n = 121) | Native T1 | 1.5 [0.8, 2.6] | 0.17 | |
| εcc | 2.3 [1.0, 5.1] | 0.049 | ||
| εls | 2.6 [1.1, 6.0] | 0.029 | ||
| Global severity score | ||||
| Base (n = 104) | Native T1 | 2.2 [ 1.1, 4.6] | 0.026 | |
| εcc | 3.3 [1.2, 3.3] | < 0.001 | ||
| εls | 2.0 [1.9, 5.7] | 0.006 | ||
| Mid (n = 132) | Native T1 | 1.3 [ 1.0, 1.9] | 0.085 | |
| εcc | 2.8 [1.7, 4.6] | < 0.001 | ||
| εls | 2.1 [1.5, 3.1] | < 0.001 | ||
| Apex (n = 105) | Native T1 | 0.9 [ 0.6, 1.5] | 0.25 | |
| εcc | 1.8 [0.9, 3.4] | 0.077 | ||
| εls | 1.8 [0.9, 3.2] | 0.085 | ||
| Global (n = 120) | Native T1 | 1.8 [ 1.1, 3.1] | 0.025 | |
| εcc | 2.6 [1.4, 4.8] | 0.002 | ||
| εls | 2.6 [1.4, 4.7] | 0.002 | ||
| FWHM | ||||
| Base (n = 103) | Native T1 | 1.4 [0.9, 2.2] | 0.19 | |
| εcc | 2.0 [1.1, 3.5] | 0.017 | ||
| εls | 1.5 [1.0, 2.2] | 0.038 | ||
| Mid (n = 114) | Native T1 | 1.1 [0.8, 1.6] | 0.58 | |
| εcc | 2.2 [1.3, 3.5] | 0.002 | ||
| εls | 1.7 [1.1, 2.5] | 0.014 | ||
| Apex (n = 103) | Native T1 | 1.0 [0.6, 1.5] | 0.85 | |
| εcc | 1.3 [0.8, 2.3] | 0.26 | ||
| εls | 0.9 [0.5, 1.6] | 0.70 | ||
| Global (n = 119) | Native T1 | 1.3 [0.8, 2.2] | 0.28 | |
| εcc | 1.4 [0.9, 2.2] | 0.19 | ||
| εls | 2.0 [1.2, 3.5] | 0.01 | ||
Bold italic values are significant for p ≤ 0.05
FWHM full width half maximum
Fig. 4.
Receiver-operator curves for noncontrast variables. Plots for combined model with native T1, Ɛcc, and εls with modest areas under the curve (AUC) for prediction of presence/absence of LGE for all CMRs (a) and in the subset with normal left ventricular ejection fraction (LVEF) (c). The best performance was for predicting high grade [global severity score (GSS) ≥ 3] vs low grade (GSS ≤ 2) for all CMRs (b)
Table 3.
Threshold performance for models incorporating native T1, ɛls, and ɛcc
| Outcome condition | Model fit equation | AUC | R2 | Threshold | Sensitivity (%) | Specificity (%) | + LR | − LR |
|---|---|---|---|---|---|---|---|---|
| Presence/absence LGE | 0.0096 * (native T1) + 0.2263 * (ɛls) + 0.538 * (ɛcc) | 0.74 | 0.21 | 0.473 | 81.1 | 42.3 | 1.4 | 0.45 |
| 0.839 | 74.3 | 69.2 | 2.4 | 0.37 | ||||
| 1.512 | 44.6 | 96.2 | 11.7 | 0.58 | ||||
| LVEF (univariate) | 0.81 | – | 56% | 56.8 | 99.9 | 568 | 0.43 | |
| High (GSS 3–4) vs low (GSS 0–2) grade LGE | 0.013 * (native T1) + 0.32 * (ɛls) + 0.059 * (ɛcc) | 0.82 | 0.38 | − 1.297 | 94.9 | 53.3 | 2.0 | 0.10 |
| − 0.559 | 76.9 | 76.7 | 3.3 | 0.30 | ||||
| 0.672 | 43.6 | 93.3 | 6.5 | 0.60 | ||||
| LVEF (univariate) | 0.87 | – | 50% | 63.8 | 93.8 | 10.3 | 0.39 |
GSS global severity score, AUC area under the curve, + LR positive likelihood ration, − LR negative likelihood ratio, LVEF left ventricular ejection fraction
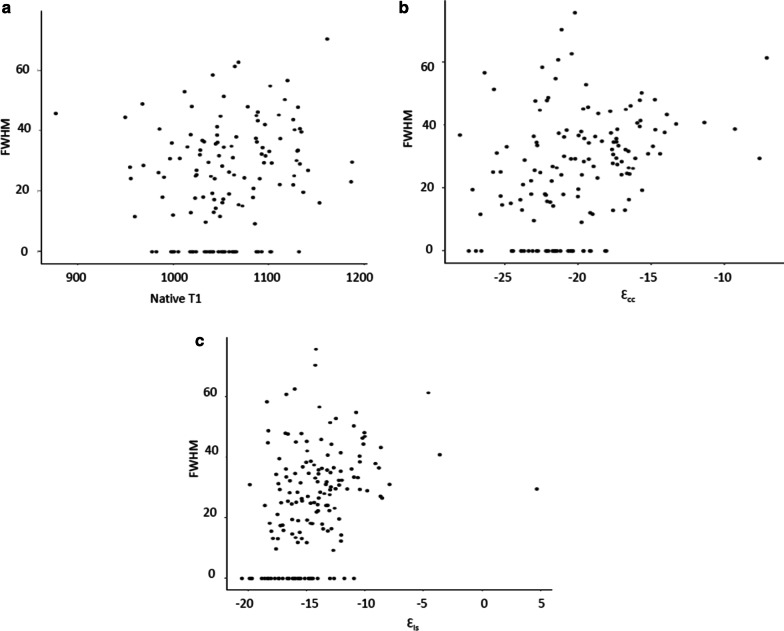
For tagged analysis, global εcc-tag was predictive of presence/absence of LGE (n = 132, OR 3.3 [1.7, 6.4], p < 0.001) and GSS (n = 131, OR 2.6 [2.2, 6.4], p < 0.001) while native T1 was predictive of GSS but not presence/absence of LGE (Additional file 3: Table S1). Scatter plots for the distribution of FWHM values versus global native T1 (A), global εcc (B), and global εls (C) are shown in Fig. 5.
Fig. 5.
Distribution of noncontrast variables as a function of full width half maximum (FWHM). Scatter plots for native T1 (a), Ɛcc (b), and εls (c) vs FWHM
A subset analysis was performed to evaluate presence of LGE in subjects with a normal LVEF. Given the smaller number of CMRs (total n = 95, 55 with LGE), the model performed poorly, with a C = 0.64 and r2 = 0.057. Figure 4c shows receiver operator curve for the model with εcc, εls, and native T1 for DMD patients with LVEF ≥ 55% (n = 95).
Models evaluating percent LGE by slice (base, mid, and apex) as predicted by strain and T1 at each slice were also evaluated. Basal and mid-LV εls, εcc, and εcc-tag were predictive of presence or absence of LGE, global severity score, and to a lesser extent FWHM (Table 2, Additional file 3: Table S1). Native T1 was only predictive in the basal slices.
For individual segments, εcc and/or εls predicted presence/absence of LGE in almost all segments assessed except for the basal anterior, mid inferolateral, and mid anterolateral segments (Table 4). Native T1 failed to significantly predict presence/absence of LGE in most segments assessed and neither native T1, εls, nor εcc were reliably predictive of segmental percent LGE (Additional file 4: Table S2). For tagged data, εcc-tag was predictive of LGE presence and extent by FWHM for mid-LV segments (Additional file 4: Table S2).
Table 4.
Models for prediction of presence/absence LGE by segment
| Segment | Factor | LGE presence/absence | p value |
|---|---|---|---|
| Odds ratio and 95% CI | |||
| Basal anterior | Native T1 | 1.2 [0.9, 1.6] | 0.33 |
| εcc | 1.3 [0.9, 1.9] | 0.21 | |
| εls | 1.1 [0.7, 1.9] | 0.72 | |
| Basal anteroseptal | Native T1 | 1.1 [0.7, 1.6] | 0.73 |
| εcc | 1.4 [0.9, 2.2] | 0.17 | |
| εls | 1.9 [1.2, 3.0] | 0.01 | |
| Basal inferoseptal | Native T1 | 1.5 [1.1, 2.1] | 0.01 |
| εcc | 3.6 [2.4, 5.5] | < 0.001 | |
| εls | 1.9 [1.2, 3.0] | 0.01 | |
| Basal inferior | Native T1 | 2.0 [1.2, 3.4] | 0.01 |
| εcc | 2.7 [1.7, 4.1] | < 0.001 | |
| εls | 1.1 [0.7, 1.6] | 0.77 | |
| Basal inferolateral | Native T1 | 2.0 [1.4, 3.3] | 0.003 |
| εcc | 2.9 [1.8, 4.7] | < 0.001 | |
| εls | 1.4 [0.9, 2.1] | 0.21 | |
| Basal anterolateral | Native T1 | 1.3 [0.8, 2.1] | 0.25 |
| εcc | 4.6 [2.8, 7.7] | < 0.001 | |
| εls | 1.8 [1.0, 3.3] | 0.042 | |
| Mid anterior | Native T1 | 0.9 [0.6, 1.2] | 0.38 |
| εcc | 2.0 [1.3, 3.2] | 0.001 | |
| εls | 1.2 [0.8 1.7] | 0.38 | |
| Mid anteroseptal | Native T1 | 1.0 [0.7, 1.5] | 0.99 |
| εcc | 1.5 [0.9, 2.6] | 0.15 | |
| εls | 1.9 [1.3, 2.8] | < 0.001 | |
| Mid inferoseptal | Native T1 | 1.3 [0.8, 2.2] | 0.26 |
| εcc | 3.2 [1.9, 5.6] | < 0.001 | |
| εls | 1.6 [1.0, 2.4] | 0.04 | |
| Mid inferior | Native T1 | 0.9 [0.5, 1.5] | 0.59 |
| εcc | 2.3 [1.3, 4.1] | 0.004 | |
| εls | 1.4 [1.0, 1.9] | 0.05 | |
| Mid inferolateral | Native T1 | 1.4 [0.8, 2.2] | 0.25 |
| εcc | 1.2 [0.5, 2.9] | 0.67 | |
| εls | 2.8 [1.9, 4.3] | < 0.001 | |
| Mid anterolateral | Native T1 | 0.9 [06, 1.4] | 0.59 |
| εcc | 1.2 [0.5, 3.2] | 0.70 | |
| εls | 1.3 [0.9, 2.0] | 0.17 |
Bold italic values are significant for p ≤ 0.05
Longitudinal assessment of LGE using native T1 and strain in individual patients
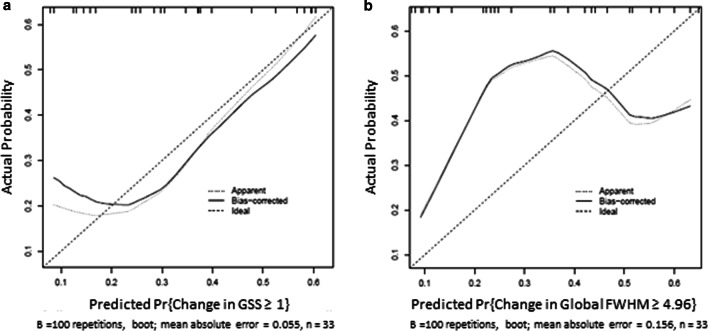
Fifty-five patients had more than one CMR and were included in the longitudinal analysis. Mean changes in follow-up time, LVEF, and CMR parameters between studies are shown in Table 5. The mean interval between the first and second CMR was 423 ± 200 days and the mean interval between the first and third CMR was 748 ± 344 days. Ordinal logistic regression modeling for change in GSS using native T1, εcc, εls, and initial global LGE demonstrated poor predictive power at the base with a C = 0.663 and r2 = 0.158 (Table 6). The model performed modestly better for native T1, Ɛcc, εls, and initial global LGE at the mid slice (C = 0.784 and r2 = 0.327) and globally (C = 0.752 and r2 = 0.288, Fig. 6a). Linear regression modeling for change in percent LGE using FWHM (Table 6) demonstrated modestly worse correlation statistics for models using the same parameters at the base (C = 0.616, r2 = 0.145), mid (C = 0.642, r2 = 0.234), and globally (C = 0.659, r2 = 0.267) (Fig. 6b). Change in FWHM was predicted by change in εcc-tag at base (C = 0.585, r2 = 0.233), mid (C = 0.617, r2 = 0.235), and globally (C = 0.636, r2 = 0.355, Table 6). There was not sufficient power to detect incremental change in GSS, although this trended toward significance (p = 0.084, data not shown).
Table 5.
Characteristics of longitudinal assessment
| N | Mean ± SD or N (%) | |
|---|---|---|
| CMR 1 to CMR 2 | ||
| Interval (days) | 55 | 423 ± 200 |
| ∆LVEF1–2 (%) | 55 | − 2.6 ± 4.7 |
| Base ∆Ɛcc1–2 | 54 | 0.7 ± 3.0 |
| Mid ∆Ɛcc1–2 | 55 | 0.7 ± 3.3 |
| Apex ∆Ɛcc1–2 | 52 | 0.6 ± 3.4 |
| ∆FWHM1–2 | 37 | 4.3 ± 14.2 |
| ∆LGE1–2 | 53 | |
| − 1 | 4 (8%) | |
| 0 | 31 (58%) | |
| 1 | 15 (28%) | |
| 2 | 2 (4%) | |
| 3 | 1 (2%) | |
| CMR 2 to CMR 3 | ||
| Interval (days) | 35 | 376 ± 26 |
| ∆LVEF2–3 (%) | 35 | − 2.4 ± 5.5 |
| Base ∆Ɛcc2–3 | 34 | 0.4 ± 3.1 |
| Mid ∆Ɛcc2–3 | 34 | 0.8 ± 3.1 |
| Apex ∆Ɛcc2–3 | 32 | 0.1 ± 3.8 |
| ∆FWHM2–3 | 27 | 3.2 ± 16.3 |
| ∆LGE2–3 | 35 | |
| − 1 | 4 (11%) | |
| 0 | 17 (49%) | |
| 1 | 11 (31%) | |
| 2 | 3 (9%) | |
| CMR 1 to CMR 3 | ||
| Interval (days) | 35 | 748 ± 344 |
| ∆LVEF1–3 (%) | 35 | − 5.1 ± 5.8 |
| Base ∆Ɛcc1–3 | 35 | 1.2 ± 3.2 |
| Mid ∆Ɛcc1–3 | 34 | 1.3 ± 3.4 |
| Apex ∆Ɛcc1–3 | 34 | 0.7 ± 4.1 |
| ∆FWHM1–3 | 23 | 9.0 ± 16.6 |
| ∆LGE1–3 | 35 | |
| − 1 | 2 (6%) | |
| 0 | 15 (43%) | |
| 1 | 13 (37%) | |
| 2 | 2 (6%) | |
| 3 | 3 (9%) | |
Table 6.
Models predicting change in LGE severity
| Slice | Factor | Odds ratio and 95% CI | p value |
|---|---|---|---|
| Change in global severity score (feature tracking) | |||
| Base (n = 27) | Native T1 | 1.2 [ 0.3, 5.0] | 0.78 |
| εcc | 2.2 [0.6, 7.7] | 0.21 | |
| εls | 1.8 [0.8, 3.9] | 0.15 | |
| LGE at CMR1 | 0.6 [0.2, 1.9] | 0.37 | |
| Mid (n = 30) | Native T1 | 1.2 [0.5, 2.7] | 0.64 |
| εcc | 0.3 [0.1, 1.1] | 0.07 | |
| εls | 3.0 [1.3, 6.7] | 0.008 | |
| LGE at CMR1 | 0.3 [0.1, 1.5] | 0.13 | |
| Global (n = 33) | Native T1 | 1.4 [0.6, 3.3] | 0.44 |
| εcc | 0.5 [0.2, 1.2] | 0.14 | |
| εls | 3.8 [ 1.0, 14.2] | 0.048 | |
| LGE at CMR1 | 0.5 [0.1, 1.7] | 0.25 | |
| Change in global severity score (myocardial tagging) | |||
| Base (n = 42) | Native T1 | 1.3 [ 0.5, 3.2] | 0.59 |
| εcc-tag | 0.8 [0.4, 1.8] | 0.56 | |
| LGE at CMR1 | 0.4 [0.1, 1.3] | 0.13 | |
| Mid (n = 44) | Native T1 | 1.5 [0.9, 2.4] | 0.11 |
| εcc-tag | 0.6 [0.3, 1.2] | 0.15 | |
| LGE at CMR1 | 0.5 [0.2, 1.6] | 0.22 | |
| Global (n = 38) | Native T1 | 1.5 [0.7, 2.9] | 0.27 |
| εcc-tag | 0.5 [0.2, 1.2] | 0.12 | |
| LGE at CMR1 | 0.3 [0.1, 1.2] | 0.10 | |
| Change in FWHM (feature tracking) | |||
| Base (n = 27) | Native T1 | 1.2 [0.3, 4.8] | 0.83 |
| εcc | 1.5 [0.4, 6.8] | 0.57 | |
| εls | 1.9 [0.7, 4.9] | 0.19 | |
| LGE at CMR1 | 0.6 [0.1, 3.7] | 0.57 | |
| Mid (n = 30) | Native T1 | 0.8 [0.4, 1.6] | 0.55 |
| εcc | 0.4 [0.1, 1.9] | 0.25 | |
| εls | 1.5 [0.5, 4.6] | 0.48 | |
| LGE at CMR1 | 0.1 [0, 1.5] | 0.10 | |
| Global (n = 33) | Native T1 | 0.9 [0.4, 2.2] | 0.84 |
| εcc | 0.7 [0.2, 2.7] | 0.61 | |
| εls | 2.0 [0.9, 4.5] | 0.10 | |
| LGE at CMR1 | 0.1 [0, 0.8] | 0.033 | |
| Change in FWHM (myocardial tagging) | |||
| Base (n = 36) | Native T1 | 1.7 [ 0.7, 4.1] | 0.26 |
| εcc-tag | 0.6 [0.5, 0.9] | 0.003 | |
| LGE at CMR1 | 0.3 [0.1, 1.6] | 0.16 | |
| Mid (n = 40) | Native T1 | 1.6 [0.7, 3.8] | 0.32 |
| εcc-tag | 0.4 [0.1, 0.9] | 0.036 | |
| LGE at CMR1 | 0.2 [0.1, 1.1] | 0.058 | |
| Global (n = 34) | Native T1 | 1.8 [0.7, 4.6] | 0.21 |
| εcc-tag | 0.2 [0.1, 0.7] | 0.01 | |
| LGE at CMR1 | 0.2 [0.03, 1.1] | 0.075 | |
Bold italic values are significant for p ≤ 0.05
Fig. 6.
Predicted vs actual probability of change in LGE. Model performance for global severity score ≥ 1 (a) and change in FWHM (b)
Changes in individual segments were also modeled. A model including change in εcc, εls, and native T1 mapping had moderate predictive power for segmental LGE changes by severity score but was less accurate for FWHM (Table 7 and Additional file 5: Table S3). Similar results were seen with tagging (Additional file 6: Table S4).
Table 7.
Models for prediction of change in LGE severity by segment
| Segment | Factor | Change in severity score | p value |
|---|---|---|---|
| Odds ratio and 95% CI | |||
| Basal anterior | Native T1 | 1.6 [1.0, 2.7] | 0.06 |
| εcc | 1.4 [0.8, 2.4] | 0.30 | |
| εls | 2.9 [1.2, 7.1] | 0.02 | |
| Basal anteroseptal | Native T1 | 3.5 [1.6, 7.6] | 0.002 |
| εcc | 1.1 [0.4, 3.1] | 0.87 | |
| εls | 0.4 [0.3, 0.8] | 0.012 | |
| Basal inferoseptal | Native T1 | 1.4 [1.0, 2.1] | 0.07 |
| εcc | 1.1 [0.3, 1.0] | 0.55 | |
| εls | 0.5 [0.3, 1.0] | 0.048 | |
| Basal inferior | Native T1 | 0.5 [0.2, 1.5] | 0.23 |
| εcc | 0.4 [0.1, 1.3] | 0.11 | |
| εls | 2.2 [1.0, 4.9] | 0.042 | |
| Basal inferolateral | Native T1 | 2.2 [0.4, 11.6] | 0.35 |
| εcc | 1.1 [0.8, 1.5] | 0.11 | |
| εls | 5.6 [1.1, 29.4] | 0.042 | |
| Basal anterolateral | Native T1 | 1.5 [1.0, 2.2] | 0.47 |
| εcc | 0.6 [0.2, 2.2] | 0.49 | |
| εls | 4.0 [0.8, 20.1] | 0.10 | |
| Mid anterior | Native T1 | 1.5 [0.6, 3.8] | 0.41 |
| εcc | 1.6 [0.6, 4.2] | 0.37 | |
| εls | 0.8 [0.5, 1.5] | 0.52 | |
| Mid anteroseptal | Native T1 | 1.0 [0.9, 1.0] | 0.07 |
| εcc | 0.4 [0.2, 1.1] | 0.06 | |
| εls | 1.5 [0.3, 7.8] | 0.63 | |
| Mid inferoseptal | Native T1 | 2.3 [0.6, 9.1] | 0.22 |
| εcc | 1.1 [0.7, 1.8] | 0.55 | |
| εls | 0.3 [0.1, 3.0] | 0.31 | |
| Mid inferior | Native T1 | 0.4 [0.2, 0.8] | 0.006 |
| εcc | 0.1 [0, 0.5] | 0.007 | |
| εls | 2.4 [0.8, 7.1] | 0.13 | |
| Mid inferolateral | Native T1 | 2.9 [1.1, 8.3] | 0.042 |
| εcc | 0.5 [0.1, 2.0] | 0.33 | |
| εls | 1.8 [0.4, 9.2] | 0.48 | |
| Mid anterolateral | Native T1 | 1.2 [0.7, 2.1] | 0.46 |
| εcc | 0.9 [0.7, 1.1] | 0.34 | |
| εls | 1.7 [0.4, 7.5] | 0.45 |
Bold italic values are significant for p ≤ 0.05
Discussion
This study presents the largest cohort of CMRs in DMD patients evaluating εcc, εls, and native T1. The primary findings of this study are: (1) modeling using pre-contrast sequences modestly predicts presence and severity of LGE for individual CMRs; (2) strain appears more predictive than native T1 in most models; (3) strain, but not native T1 or initial global LGE, predicts change in severity of LGE estimated by GSS (for feature tracking) and FWHM (for myocardial tagging); (4) Non-contrast sequences are not a substitute for LGE imaging in DMD. While strain was statistically predictive of most outcome variables, the performance of strain was only modest, and native T1 was not predictive in most models. The performance of most non-contrast models evaluated did not meet thresholds for routine clinical use, emphasizing the importance of LGE in the DMD population.
Native T1 mapping has shown promise as a tool for detecting myocardial fibrosis in dilated cardiomyopathy as assessed by LGE and histologically [33]. Given the regionality of fibrosis observed in DMD cardiomyopathy, the utility of this sequence may be in selected segments rather than as a global predictor. Apical slices are susceptible to errors from partial volume averaging and also have less fibrosis than basal and mid-LV slices in DMD boys. Global values were thus calculated excluding apical slices. Recently, Olivieri and colleagues found that native T1 and ECV in the lateral wall were increased in DMD boys with LGE compared to those without LGE, though there was no significant difference in the septum [34]. They also found that the saturation recovery single shot acquisition (SASHA) technique was better at discriminating between disease states (control, DMD no LGE, and DMD with LGE) than MOLLI; however, modeling suggested that MOLLI could also distinguish between DMD boys with and without LGE. Studies suggest that native T1 values of myocardium and, to a lesser extent blood pool, remain relatively stable in healthy adults [35] and those with arrhythmias [36]. However, systematic longitudinal assessment of native T1 reproducibility has not been well characterized in dilated cardiomyopathies. There are many factors that may have affected the association of T1 maps with LGE in the DMD patient population. Fatty infiltration has been well-described in skeletal muscle and may be present with progressive myocardial disease. This could lower native T1– thus, T1 values may depend on the ratio of fibrosis to fat in LGE. Indeed, a recent study suggested native T1 decreases with worsening LGE in DMD boys, which the authors hypothesized was secondary to fatty infiltration [37]. DMD boys also may have lower image quality as they age, as many DMD boys have progressive respiratory weakness, limiting ability for adequate breath-holding. This limitation should be corrected in most cases with the motion correction and we excluded patients with poor map quality. Positioning and image quality may lead to scan-to-scan variation in native T1 maps. This may lead to inaccurate estimation of the change in native T1. Finally, while LGE demonstrates replacement fibrosis, native T1 is increased with both replacement and diffuse fibrosis; in addition, native T1 detects abnormalities in both myocytes and the extracellular matrix. Taken together, these factors may contribute to the poor performance of our models when using native T1 as one of the predictors for global severity as assessed by LGE.
By contrast, strain is a more functional assessment and areas demonstrating abnormal strain patterns secondary to fibrotic changes would be expected to progress as the pathological process progresses. Good correlation with εcc and myocardial fibrosis has been previously demonstrated based on histopathology and CMR assessment with LGE and ECV in adult patients with aortic stenosis [38] and hypertrophic cardiomyopathies [39]. Our data suggest that this holds true for dilated cardiomyopathy in DMD boys, with strain predicting both change in presence/absence and change in LGE extent in most of our models. Of note, while strain predicts change in extent of LGE, model performance is suboptimal (Fig. 6). Further, univariate analysis demonstrates that LVEF is at least as good as our multivariate models with native T1 and strain in predicting presence or absence of LGE in DMD patients with normal function (Additional file 1: Figure S1). While LVEF alone predicts the presence of LGE with high specificity (although less robust sensitivity), this is not as useful in clinical practice as the population of interest is DMD patients with normal or stable LVEF and progression of LGE. Interestingly, our model performs better in terms of predictive power for all DMD boys compared to the subset with normal LVEF (≥ 55%) and our models have better negative likelihood ratios than LVEF alone (Table 3). This is consistent with studies in adult populations with chronic heart failure, where addition of strain to LVEF had modest but significant improvements in AUCs [40]. This suggests a very limited role for strain, where one could consider using it only in those DMD boys who have an absolute contraindication to gadolinium or in whom intravenous access cannot be obtained. Given the suboptimal results of most models, the authors cannot recommend routine use of non-contrast CMR in place of LGE in patients without a contraindication to gadolinium. Despite the safety concerns associated with contrast, DMD subjects will likely continue to require conservative administration of gadolinium contrast for appropriate clinical management.
It is important to note the segmental distribution of the predictive models using native T1 and strain. Unlike many other forms of cardiomyopathy, the fibrosis seen in DMD dilated cardiomyopathy tends to be patchy and found in the subepicardial region, often in one or more segments of the lateral free wall. Segmental differences in native T1 and strain are more likely to be predictive in areas where LGE is usually found in DMD patients. Thus, there may be some benefit to monitoring for changes in segmental parameters even in patients with normal LVEF and no previous LGE. Strain calculated using either feature tracking or myocardial tagging was predictive of LGE presence and extent with similar efficacy. It should be noted that HARP analyzes mid-myocardial strain while feature tracking analyzes endocardial strain. There is debate about which one is more relevant clinically, but we suspect that the difference is a result of the underlying software differences. Apical LGE was better predicted by εcc-tag, however, this likely is of less clinical importance given that the apex in DMD is relatively spared until late in the disease. It is still unclear what outcome measures are best used for DMD cardiomyopathy. LVEF and LGE have been the standard because clinicians have the most experience with their use and interpretation [11]. It is possible that more advanced methods, such as native T1, ECV, and strain, will supplant LVEF and LGE for assessment of DMD cardiomyopathy severity and prediction of progression, but this study was not designed to determine the optimal CMR biomarkers, only whether non-contrast sequences can predict LGE.
Emphasizing the importance of limiting contrast exposure, a recent study demonstrated no difference in medical management seen with traditional versus conservative contrast administration in DMD patients [41]. Of note, the conservative contrast group still received contrast 84% of the time, suggesting that conservative contrast administration may be just the first step in decreasing cumulative contrast load in DMD. While our institution has been using conservative gadolinium administration for the past 3 years, non-contrast sequences, particularly εls and Ɛcc, have potential to help in further reducing contrast administration in this patient population.
Given the substantial mobility challenges and other morbidities, such as contractures and severe scoliosis, seen in DMD patients, reduction in scanner time is an important consideration for patient comfort as well as patient compliance for breath holding in order to obtain the best quality studies. Based on our institutional data, the average total scan time was 39.6 min, with 10 min attributed to post-contrast imaging from sequences that would not have been used if contrast was not administered. Thus, eliminating contrast administration would allow for sub-30 min studies in most cases, which is an important consideration for this patient population in particular, but may also be important in centers were scanner access is limited.
Limitations
This study is limited by the fact that only 33% (18 of 55 patients) had a change in GSS ≥ 1; a larger change may have improved model performance. Additionally, 70% of patients had LGE severity score of at least 1 on their initial scan. This limited our ability to do subset analysis with sufficient power comparing the severity score 0 group alone with those ≥ 1. As discussed above, native T1 may not be completely linearly related to LGE extent and this may bias the data toward non-significance when there actually is a detectable difference for larger changes in LGE severity. Additionally, the variation in native T1 values is inversely related to flip angle, with lower flip angles having higher standard deviations between measurements [36]. We chose to use MOLLI due to its wide availability, which potentially make our findings more generalizable. However, it is possible that other methods of T1 mapping, such as SASHA, would provide a better prediction of LGE. There were some patients that were excluded because we were not able to obtain IV access in order to administer contrast and a very small minority had to stop early due to poor endurance or discomfort. These patients were not included if they did not complete the protocol; as this was a small minority of patients, we do not believe this led to any bias in the results. It should be noted that any patient with a heart rate > 90 bpm used the high heart rate version of the T1 map. In our data set, 72% of the CMRs were performed in the setting of heart rates > 90 bpm. While this sequence increases the voxel size by decreasing the matrix size, this correction significantly decreases the sensitivity of MOLLI to heart rates at longer T1 times. Our prior work demonstrated no significant effect of HR on T1 times using these parameters [21]. Further investigation with a larger, multicenter cohort may allow for better resolution of detectible differences in native T1 in patients who have small changes in the extent of LGE.
Most of the patients assessed were on steroids or cardiovascular medications. While medications may temper the progression of LGE, which could certainly increase the number of patients with no change in LGE on longitudinal assessment, therapies are unlikely to substantially change the predictive potential of these sequences. Despite these therapies, there does appear to be a relationship with mortality. It is important to assess prediction in a clinically relevant setting, and all of these medications represent standard therapies at most institutions.
Finally, another limitation of this analysis is that the models will require validation secondary to the large number of multiple comparisons.
Conclusions
In this cohort of DMD boys, models using pre-contrast sequences are predictive of presence and severity of LGE for a particular CMR using both feature tracking and myocardial tagging. For longitudinal assessment for individual patients, εcc-tag was superior for identification of change in severity by FWHM and εls was superior for change in GSS. Although these models were predictive, they were only modestly so, suggesting that non-contrast studies cannot currently replace LGE imaging. Given the critical clinical data provided by LGE, we suggest that DMD subjects should continue to receive gadolinium contrast using a conservative protocol for clinical management. Non-contrast imaging should only be used to estimate LGE when contrast cannot be administered (renal failure or no intravenous access) or if total time must be decreased due to physical limitations.
Supplementary Information
Additional file 1: Figure S1. Univariate Prediction Models for LVEF, LVESVin, and LVEDVin. Models demonstrate modest areas under the curve (AUC) for prediction of presence/absence of LGE for LVEF (A), LVESVin (B) and poor performance for LVEDVin (C). LVEF does well for predicting high grade (GSS ≥ 3) vs low grade (GSS ≤ 2) LGE (D), with less robust performance by LVESVin (E) and LVEDVin (F).
Additional file 2: Figure S2. Analysis of missing data points in longitudinal cohort. The variable with the highest proportion of missing values was change in basal ɛls (A). Combination analysis was performed with blue squares indicating observed data points and red squares indicating missing data (B). This demonstrated the majority of CMRs had all data points (indicated by the bar graph to the right of the matrix), with the next most common being missing only change in basal ɛls.
Additional file 3: Table S1. Models for prediction of presence and severity of LGE (myocardial tagging).
Additional file 4: Table S2. Models for prediction of presence/absence LGE and FWHM by segment (myocardial tagging).
Additional file 5: Table S3. Models for prediction of FWHM and change in FWHM by segment (feature tracking).
Additional file 6: Table S4. Models for prediction of change in LGE severity by segment (myocardial tagging).
Acknowledgements
The authors would like to acknowledge the developers of the pulse sequence and reconstruction software: Bruce Spottiswoode, Andreas Greiser, Hui Xue, Christopher Glielmi, Shivraman Giri, and Randall Kroeker.
Abbreviations
- εcc
Circumferential strain
- εcc-tag
Circumferential strain calculated from myocardial tagging
- εls
Longitudinal strain
- bSSFP
Balanced steady state free precession
- CMR
Cardiovascular magnetic resonance
- DMD
Duchenne muscular dystrophy
- ECG
Electrocardiogram
- ECV
Extracellular volume fraction
- FWHM
Full width half maximum
- GSS
Global severity score
- HARP
Harmonic phase magnetic resonance
- LGE
Late gadolinium enhancement
- LV
Left ventricle/left ventricular
- LVEDV
Left ventricular end-diastolic volume
- LVEDVI
Left ventricular end-diastolic volume index
- LVEF
Left ventricular ejection fraction
- LVESV
Left ventricular end-systolic volume
- LVESVI
Left ventricular end-systolic volume index
- MOLLI
Modified Look-Locker inversion recovery
- PSIR
Phase sensitive inversion recovery
- RV
Right ventricle/right ventricular
- SASHA
Saturation recovery single-shot acquisition
Authors’ contributions
FR helped with design study, analyzed and interpreted the data, and drafted the manuscript. JS and DP designed the study and supervised the imaging studies to ensure appropriate quality for analysis. JS also aided in data analyzation. KGD preformed contours and analysis for LVEF, T1 maps, and strain. KC helped with patient recruitment and clinical data acquisition. LH helped design the study. MX and JCS provided aid with biostatistical analysis. All authors contributed to the manuscript. All authors read and approved the final manuscript.
Funding
Research reported in this publication was supported by the National Heart, Lung, and Blood Institute of the National Institutes of Health under Award Number K23HL123938 and R56HL141248 (Bethesda, Maryland, USA) (JS), F32HL137394 (Bethesda, MD) (FR), and K12HD087023 (Bethesda, Maryland, USA), (FR).
Other support provided by American Heart Association Grant 13CRP14530007 (JS) (Dallas, Texas, USA), the Fighting Duchenne Foundation and the Fight DMD/Jonah & Emory Discovery Grant (Nashville, Tennessee, USA) (LM), and United States Food and Drug Administration Grant R01FD006649 (JS).
Additional project support obtained through CTSA award No. UL1 TR002243 from the National Center for Advancing Translational Sciences. The content of this work is solely the responsibility of the authors and does not necessarily represent official views of the National Center for Advancing Translational Sciences or the National Institutes of Health.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
This single-center, retrospective, observational study was approved by the Vanderbilt University Institutional Review Board. Participants older than 18 years gave written consent. For children less than 18 years, consent was obtained from legal guardians.
Consent for publication
Not applicable.
Competing interests
The authors have no relationship with industry or competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Dooley J, Gordon KE, Dodds L, MacSween J. Duchenne muscular dystrophy: a 30-year population-based incidence study. Clin Pediatr (Phila) 2010;49:177–9. doi: 10.1177/0009922809347777. [DOI] [PubMed] [Google Scholar]
- 2.Nigro G, Comi LI, Politano L, Bain RJ. The incidence and evolution of cardiomyopathy in Duchenne muscular dystrophy. Int J Cardiol. 1990;26:271–7. doi: 10.1016/0167-5273(90)90082-G. [DOI] [PubMed] [Google Scholar]
- 3.Duboc D, Meune C, Pierre B, et al. Perindopril preventive treatment on mortality in Duchenne muscular dystrophy: 10 years’ follow-up. Am Heart J. 2007;154:596–602. doi: 10.1016/j.ahj.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 4.Raman SV, Hor KN, Mazur W, et al. Eplerenone for early cardiomyopathy in Duchenne muscular dystrophy: a randomised, double-blind, placebo-controlled trial. Lancet Neurol. 2015;14:153–61. doi: 10.1016/S1474-4422(14)70318-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Power LC, O’Grady GL, Hornung TS, Jefferies C, Gusso S, Hofman PL. Imaging the heart to detect cardiomyopathy in Duchenne muscular dystrophy: a review. Neuromuscul Disord. 2018;28:717–730. doi: 10.1016/j.nmd.2018.05.011. [DOI] [PubMed] [Google Scholar]
- 6.Soslow JH, Xu M, Slaughter JC, et al. Evaluation of echocardiographic measures of left ventricular function in patients with Duchenne muscular dystrophy: assessment of reproducibility and comparison to cardiac magnetic resonance imaging. J Am Soc Echocardiogr. 2016;29:983–91. doi: 10.1016/j.echo.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Silva MC, Meira ZM, Gurgel Giannetti J, et al. Myocardial delayed enhancement by magnetic resonance imaging in patients with muscular dystrophy. J Am Coll Cardiol. 2007;49:1874–9. doi: 10.1016/j.jacc.2006.10.078. [DOI] [PubMed] [Google Scholar]
- 8.Puchalski MD, Williams RV, Askovich B, et al. Late gadolinium enhancement: precursor to cardiomyopathy in Duchenne muscular dystrophy? Int J Cardiovasc Imaging. 2009;25:57–63. doi: 10.1007/s10554-008-9352-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kieny P, Chollet S, Delalande P, et al. Evolution of life expectancy of patients with Duchenne muscular dystrophy at AFM Yolaine de Kepper centre between 1981 and 2011. Ann Phys Rehabil Med. 2013;56:443–54. doi: 10.1016/j.rehab.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 10.Menon SC, Etheridge SP, Liesemer KN, et al. Predictive value of myocardial delayed enhancement in Duchenne muscular dystrophy. Pediatr Cardiol. 2014;35:1279–85. doi: 10.1007/s00246-014-0929-z. [DOI] [PubMed] [Google Scholar]
- 11.Tandon A, Villa CR, Hor KN, et al. Myocardial fibrosis burden predicts left ventricular ejection fraction and is associated with age and steroid treatment duration in duchenne muscular dystrophy. J Am Heart Assoc. 2015;4:e001338. doi: 10.1161/JAHA.114.001338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Motoki T, Shimizu-Motohashi Y, Saito I, et al. Renal dysfunction can occur in advanced-stage Duchenne muscular dystrophy. Muscle Nerve. 2020;61:192–7. doi: 10.1002/mus.26757. [DOI] [PubMed] [Google Scholar]
- 13.Olchowy C, Maciag EJ, Sanchez-Montanez A, Olchowy A, Delgado I, Vazquez E. Measurements of signal intensity of globus pallidus and dentate nucleus suggest different deposition characteristics of macrocyclic GBCAs in children. PLoS ONE. 2018;13:e0208589. doi: 10.1371/journal.pone.0208589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kellman P, Wilson JR, Xue H, Ugander M, Arai AE. Extracellular volume fraction mapping in the myocardium, part 1: evaluation of an automated method. J Cardiovasc Magn Reson. 2012;14:63. doi: 10.1186/1532-429X-14-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.aus dem Siepen F, Buss SJ, Messroghli D, et al. T1 mapping in dilated cardiomyopathy with cardiac magnetic resonance: quantification of diffuse myocardial fibrosis and comparison with endomyocardial biopsy. Eur Heart J Cardiovasc Imaging. 2015;16:210–6. doi: 10.1093/ehjci/jeu183. [DOI] [PubMed] [Google Scholar]
- 16.Fontana M, White SK, Banypersad SM, et al. Comparison of T1 mapping techniques for ECV quantification. Histological validation and reproducibility of ShMOLLI versus multibreath-hold T1 quantification equilibrium contrast CMR. J Cardiovasc Magn Reson. 2012;14:88. doi: 10.1186/1532-429X-14-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dass S, Suttie JJ, Piechnik SK, et al. Myocardial tissue characterization using magnetic resonance noncontrast t1 mapping in hypertrophic and dilated cardiomyopathy. Circ Cardiovasc Imaging. 2012;5:726–33. doi: 10.1161/CIRCIMAGING.112.976738. [DOI] [PubMed] [Google Scholar]
- 18.Ferreira VM, Piechnik SK, Dall’Armellina E, et al. T(1) mapping for the diagnosis of acute myocarditis using CMR: comparison to T2-weighted and late gadolinium enhanced imaging. JACC Cardiovasc Imaging. 2013;6:1048–58. doi: 10.1016/j.jcmg.2013.03.008. [DOI] [PubMed] [Google Scholar]
- 19.Malek LA, Werys K, Klopotowski M, et al. Native T1-mapping for non-contrast assessment of myocardial fibrosis in patients with hypertrophic cardiomyopathy-comparison with late enhancement quantification. Magn Reson Imaging. 2015;33:718–24. doi: 10.1016/j.mri.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 20.Zhou H, Lin X, Fang L, et al. Characterization of compacted myocardial abnormalities by cardiac magnetic resonance with native T1 mapping in left ventricular non-compaction patients—a comparison with late gadolinium enhancement. Circulation journal: official journal of the Japanese Circulation Society. 2016;80:1210–6. doi: 10.1253/circj.CJ-15-1269. [DOI] [PubMed] [Google Scholar]
- 21.Soslow JH, Damon SM, Crum K, et al. Increased myocardial native T1 and extracellular volume in patients with Duchenne muscular dystrophy. J Cardiovasc Magn Reson. 2016;18:5. doi: 10.1186/s12968-016-0224-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Erley J, Genovese D, Tapaskar N, et al. Echocardiography and cardiovascular magnetic resonance based evaluation of myocardial strain and relationship with late gadolinium enhancement. J Cardiovasc Magn Reson. 2019;21:46. doi: 10.1186/s12968-019-0559-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schulz-Menger J, Bluemke DA, Bremerich J, et al. Standardized image interpretation and post processing in cardiovascular magnetic resonance: Society for Cardiovascular Magnetic Resonance (SCMR) board of trustees task force on standardized post processing. J Cardiovasc Magn Reson. 2013;15:35. doi: 10.1186/1532-429X-15-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Messroghli DR, Radjenovic A, Kozerke S, Higgins DM, Sivananthan MU, Ridgway JP. Modified Look-Locker inversion recovery (MOLLI) for high-resolution T1 mapping of the heart. Magn Resonan Med. 2004;52:141–6. doi: 10.1002/mrm.20110. [DOI] [PubMed] [Google Scholar]
- 25.Kellman P, Hansen MS. T1-mapping in the heart: accuracy and precision. J Cardiovasc Magn Reson. 2014;16:2. doi: 10.1186/1532-429X-16-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xue H, Shah S, Greiser A, et al. Motion correction for myocardial T1 mapping using image registration with synthetic image estimation. Magn Resonan Med. 2012;67:1644–55. doi: 10.1002/mrm.23153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soslow JH, Damon BM, Saville BR, et al. Evaluation of post-contrast myocardial t1 in duchenne muscular dystrophy using cardiac magnetic resonance imaging. Pediatr Cardiol. 2015;36:49–56. doi: 10.1007/s00246-014-0963-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soslow JH, Xu M, Slaughter JC, et al. The role of matrix metalloproteinases and tissue inhibitors of metalloproteinases in Duchenne muscular dystrophy cardiomyopathy. J Card Fail. 2019;25:259–67. doi: 10.1016/j.cardfail.2019.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simpson SA, Field SL, Xu M, Saville BR, Parra DA, Soslow JH. Effect of weight extremes on ventricular volumes and myocardial strain in repaired tetralogy of fallot as measured by CMR. Pediatr Cardiol. 2017;39:575–584. doi: 10.1007/s00246-017-1793-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cerqueira MD, Weissman NJ, Dilsizian V, et al. Standardized myocardial segmentation and nomenclature for tomographic imaging of the heart. A statement for healthcare professionals from the Cardiac Imaging Committee of the Council on Clinical Cardiology of the American Heart Association. Circulation. 2002;105:539–42. doi: 10.1161/hc0402.102975. [DOI] [PubMed] [Google Scholar]
- 31.Moon JC, Messroghli DR, Kellman P, et al. Myocardial T1 mapping and extracellular volume quantification: a Society for Cardiovascular Magnetic Resonance (SCMR) and CMR Working Group of the European Society of Cardiology consensus statement. J Cardiovasc Magn Reson. 2013;15:92. doi: 10.1186/1532-429X-15-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)—a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;42:377–81. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shah RV, Kato S, Roujol S, et al. Native myocardial T1 as a biomarker of cardiac structure in non-ischemic cardiomyopathy. Am J Cardiol. 2016;117:282–8. doi: 10.1016/j.amjcard.2015.10.046. [DOI] [PubMed] [Google Scholar]
- 34.Olivieri LJ, Kellman P, McCarter RJ, Cross RR, Hansen MS, Spurney CF. Native T1 values identify myocardial changes and stratify disease severity in patients with Duchenne muscular dystrophy. J Cardiovasc Magn Reson. 2016;18:72. doi: 10.1186/s12968-016-0292-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aus dem Siepen F, Baumgartner C, Muller-Henessen M, et al. Variability of cardiovascular magnetic resonance (CMR) T1 mapping parameters in healthy volunteers during long-term follow-up. Open Heart. 2018;5:e000717. doi: 10.1136/openhrt-2017-000717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hong K, Collins J, Lee DC, et al. Optimized AIR and investigational MOLLI cardiac T1 mapping pulse sequences produce similar intra-scan repeatability in patients at 3T. NMR Biomed. 2016;29:1454–63. doi: 10.1002/nbm.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lang SM, Alsaied T, Khoury PR, Ryan TD, Taylor MD. Variations in native T1 values in patients with Duchenne muscular dystrophy with and without late gadolinium enhancement. Int J Cardiovasc Imaging. 2020 doi: 10.1007/s10554-020-02031-z. [DOI] [PubMed] [Google Scholar]
- 38.Podlesnikar T, Delgado V, Bax JJ. Cardiovascular magnetic resonance imaging to assess myocardial fibrosis in valvular heart disease. Int J Cardiovasc Imaging. 2018;34:97–112. doi: 10.1007/s10554-017-1195-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu R, An DA, Shi RY, et al. Myocardial fibrosis evaluated by diffusion-weighted imaging and its relationship to 3D contractile function in patients with hypertrophic cardiomyopathy. J Magn Resonan Imaging. 2018;48:1139–46. doi: 10.1002/jmri.26016. [DOI] [PubMed] [Google Scholar]
- 40.Zhang KW, French B, May Khan A, et al. Strain improves risk prediction beyond ejection fraction in chronic systolic heart failure. J Am Heart Assoc. 2014;3:e000550. doi: 10.1161/JAHA.113.000550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lang SM, Alsaied T, Moore RA, Rattan M, Ryan TD, Taylor MD. Conservative gadolinium administration to patients with Duchenne muscular dystrophy: decreasing exposure, cost, and time, without change in medical management. Int J Cardiovasc Imaging. 2019;35:2213–2219. doi: 10.1007/s10554-019-01670-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Univariate Prediction Models for LVEF, LVESVin, and LVEDVin. Models demonstrate modest areas under the curve (AUC) for prediction of presence/absence of LGE for LVEF (A), LVESVin (B) and poor performance for LVEDVin (C). LVEF does well for predicting high grade (GSS ≥ 3) vs low grade (GSS ≤ 2) LGE (D), with less robust performance by LVESVin (E) and LVEDVin (F).
Additional file 2: Figure S2. Analysis of missing data points in longitudinal cohort. The variable with the highest proportion of missing values was change in basal ɛls (A). Combination analysis was performed with blue squares indicating observed data points and red squares indicating missing data (B). This demonstrated the majority of CMRs had all data points (indicated by the bar graph to the right of the matrix), with the next most common being missing only change in basal ɛls.
Additional file 3: Table S1. Models for prediction of presence and severity of LGE (myocardial tagging).
Additional file 4: Table S2. Models for prediction of presence/absence LGE and FWHM by segment (myocardial tagging).
Additional file 5: Table S3. Models for prediction of FWHM and change in FWHM by segment (feature tracking).
Additional file 6: Table S4. Models for prediction of change in LGE severity by segment (myocardial tagging).
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.