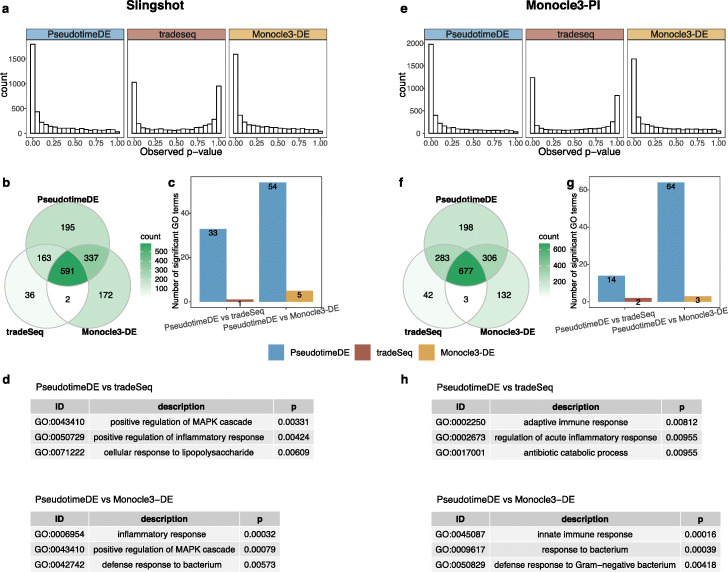
Fig. 4.
Application of PseudotimeDE, tradeSeq, and Monocle3-DE to the LPS-dendritic cell dataset. Left panels a–d are based on pseudotime inferred by Slingshot; right panels e–h are based on pseudotime inferred by Monocle3-PI. a, e Histograms of all genes’ p-values by the three DE methods. The bimodal distributions of tradeSeq’s p-values suggest a violation of the requirement that p-values follow the Uniform [0,1] distribution under the null hypothesis. b, f Venn plots showing the overlaps of the significant DE genes (BH adjusted- p≤0.01) identified by the three DE methods. PseudotimeDE’s DE genes almost include tradeSeq’s. c, g Numbers of GO terms enriched (p<0.01) in the significant DE genes specifically found by PseudotimeDE or tradeSeq/Monocle3-DE in pairwise comparisons between PseudotimeDE and tradeSeq/Monocle3-DE in b and f. Many more GO terms are enriched in the PseudotimeDE-specific DE genes than in the tradeSeq- or Monocle3-DE-specific ones. d, h Example GO terms enriched in the Pseudotime-specific DE genes in c and g. Many of these terms are related to LPS, immune process, and defense to bacterium