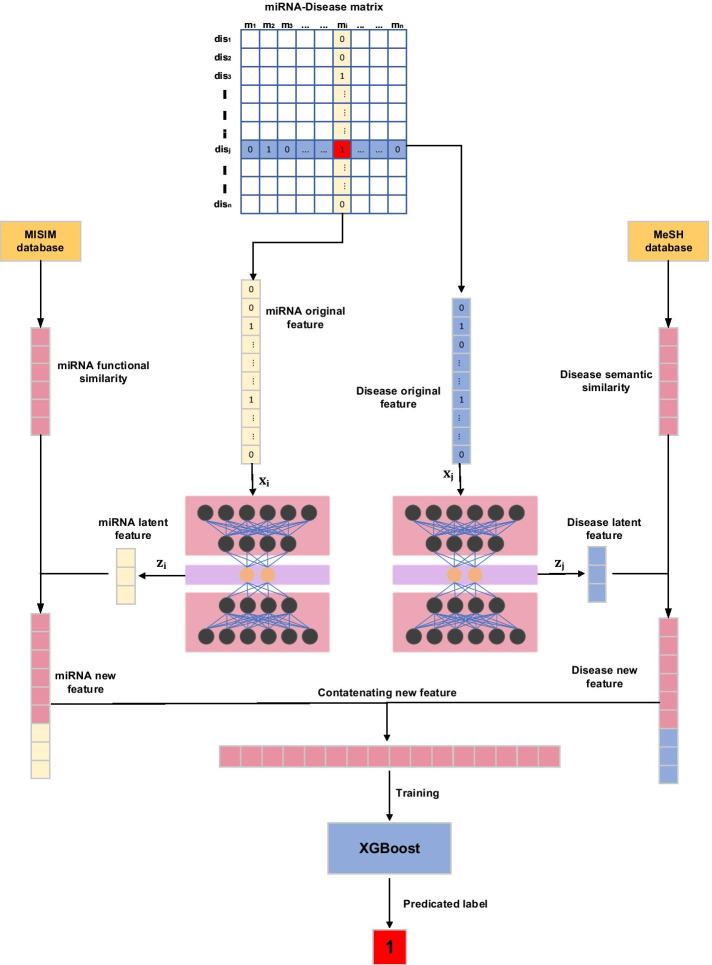
Fig. 5.
Overview of our proposed SMALF method for predicting miRNA-disease assoications.SMALF consists of four parts: Step1, we decompose miRNA-disease matrix Y into miRNA original feature M and disease original feature . Step2, we utilize stacked autoencoders to learn the latent features of miRNAs and diseases from the original feature M and . Step3,Integrating miRNA functional similarity, miRNA latent feature, disease semantic similarity, and disease latent feature generates the feature vector representing miRNA-disease. Step4, the XGBoost algorithm is employed to predict the miRNA-disease associations