Fig. 1.
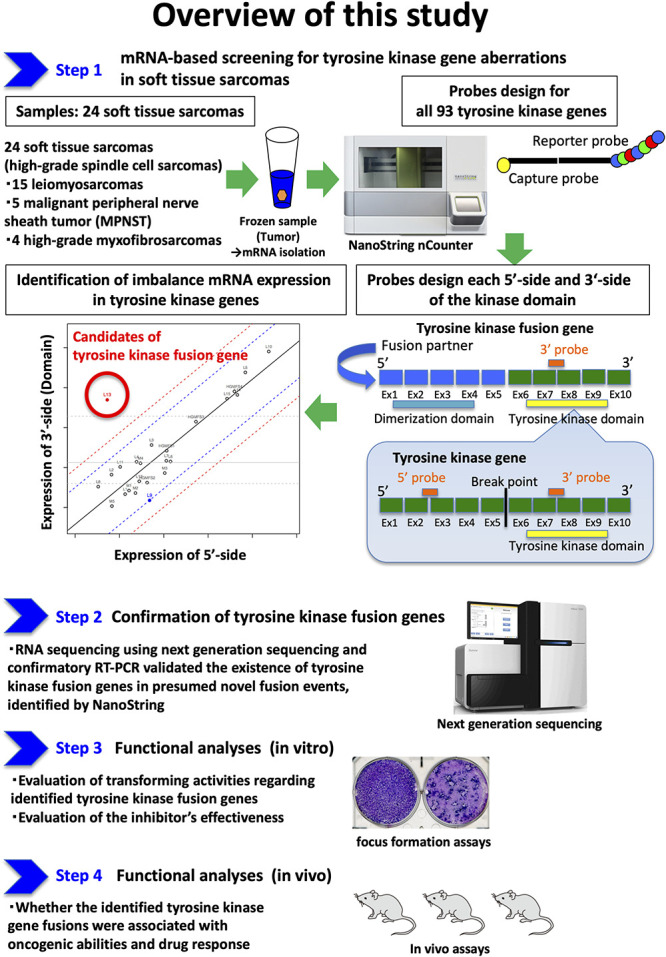
Step 1: To identify tyrosine kinase fusion genes, NanoString-based transcriptome assay analyzed 24 tumor samples with STSs including 15 leiomyosarcomas, five with MPNSTs, and four with high-grade myxofibrosarcomas. This assay screened the expression balances regarding transcripts of 90 tyrosine kinases at two points: the 5’ and 3’ endpoints of the kinase domain. Step 2: RNA sequencing using next generation sequencing and confirmatory reverse transcription-polymerase chain reaction (RT-PCR) validated whether NanoString identified existing tyrosine kinase fusion genes in presumed novel fusion events. Step 3: Functional in vitro analyses were performed to elucidate whether the identified tyrosine kinase gene fusions were associated with oncogenic abilities. Step 4: Functional analyses consisting of in vivo assays were also performed to elucidate whether the identified tyrosine kinase gene fusions were associated with oncogenic abilities and drug responses.
