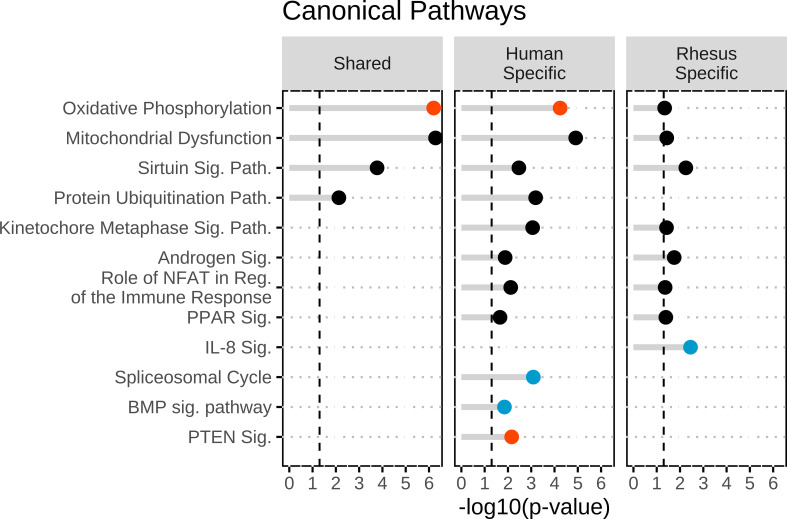
Figure 1.
Top canonical pathways identified within the shared DEGs, human-specific DEGs, and rhesus-specific DEGs groups. Canonical pathways were identified in ingenuity pathway analysis and then were graphed in R. Significance was set at a −log10(P value) of 1.3 or greater as indicated by the vertical dashed lines and all entries were found to be significant. The red dots indicate activation of the canonical pathway with a z > 1.96. The blue dots indicated inhibition of the canonical pathway with a z < −1.96. The black dots indicate no significant z-score to determine directionally of the canonical pathways. DEGs, differentially expressed genes.