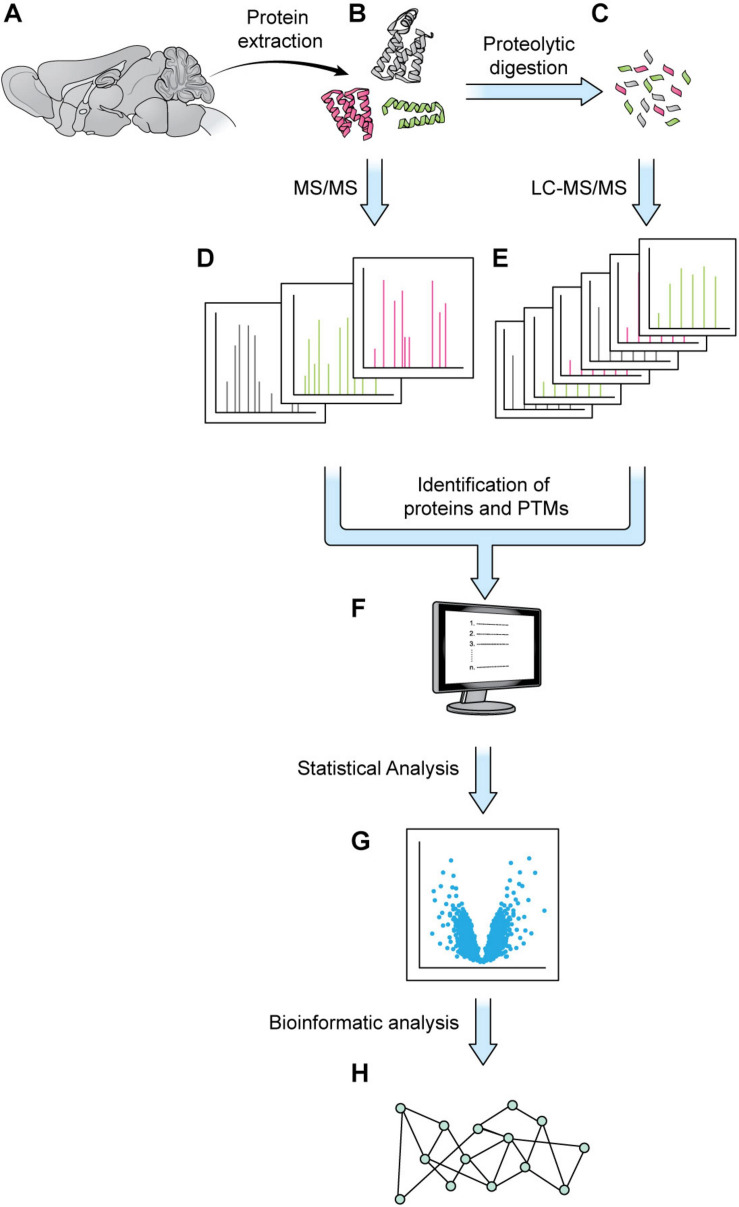
FIGURE 1.
Generic mass spectrometry-based proteomics workflow. Proteins are extracted from a biological sample. With the bottom-up approach, proteins are digested into shorter peptides, then used for LC-MS/MS data acquisition (steps A–C,E, without step D). With the top-down approach, the intact protein is subjected to MS/MS analysis without digestion (steps A,B,D, no steps C,E). MS1 and MS2 data are processed by a database search or a de novo sequencing algorithm to obtain protein sequence and PTM status (F). The abundance of MS1 and/or MSn signal can be employed for protein quantitation and statistical analysis (G). Various bioinformatics tools such as GO analysis or pathway analysis can be used to extract biologically relevant information from proteins of interest discovered in proteomics analysis (H).