Figure 2. PHGDH variants identified in MacTel patients disrupted enzyme function.
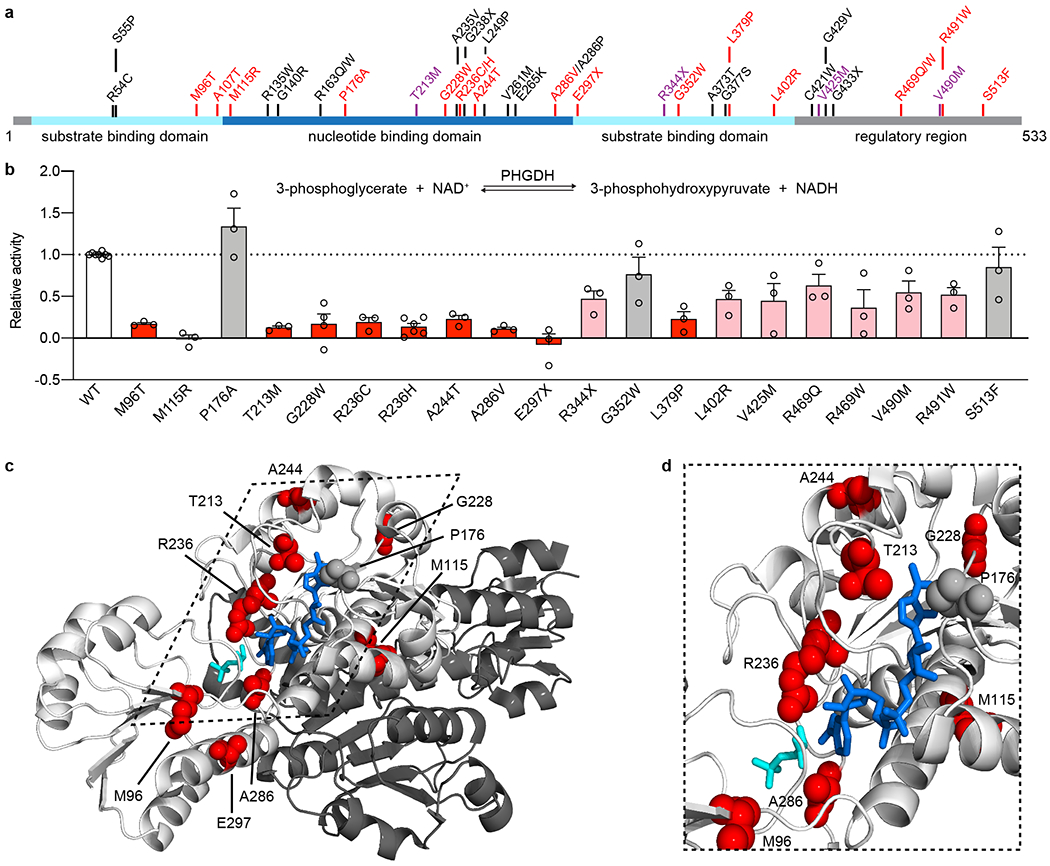
(a) Schematic of human PHGDH domain structure indicating identified MacTel variants (red), known PHGDH-deficiency/NLS1 variants (black) 11–14,19,23,38–42 and variants found in both groups (purple). (b) Relative activity of PHGDH variants compared to wild type (WT) PHGDH. Assayed activity was corrected for endogenous PHGDH by subtracting the activity of the empty vector and then normalized to the protein expression of the exogenous variant. Data for variants that retain less than 25% activity are shown in red, between 25-75% activity are shown in pink, and more than 75% activity are shown in grey. Data are shown as the mean +/− SEM, n = 3 independent experiments. A schematic of the PHGDH enzymatic reaction is shown as an inset at the top of the graph. (c) Structural representation of the location of MacTel variants on the available partial PHGDH structure. Chain A is shown in white and chain B is shown in dark grey, with substrates and variants only shown on chain A. The substrates NADH and 3PG are shown in dark blue and cyan. MacTel variants are shown in red or grey according to their activity in panel b. (d) Zoomed in and rotated view of the PHGDH active site with chain B omitted for clarity.
