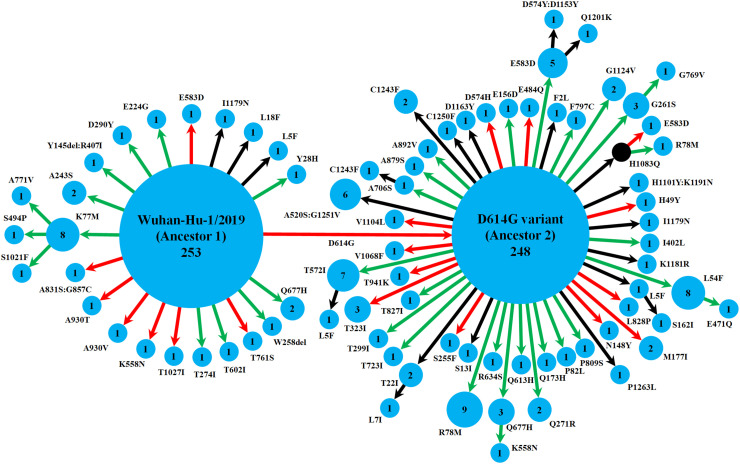
Fig. 1.
Schematic representation of the diversity of spike protein variants circulating in India, using maximum likelihood-based phylogeny reconstruction. Each node represents a specific spike protein variant, while the node-size and the number inside depict the frequency of that variant. The red or green color of each arrow indicates the higher or lower stability index respectively of the S-R complex for each variant than the major ancestral variant it emerged from (either Ancestor 1 or Ancestor 2). The black arrows lead to the variants for which the docking scores could not be determined either because of the presence of at least one variation outside the available template region for docking (Walls et al., 2020; Wrapp et al., 2020), or due to non-existing isolate in the lone hypothetical node with H1083Q mutation denoted by black color. This black node signifies a variant with no available isolate in our dataset, while it gives rise to two derived variants, H1083Q:R78M and H1083Q:E583D, for which representative isolates were available.