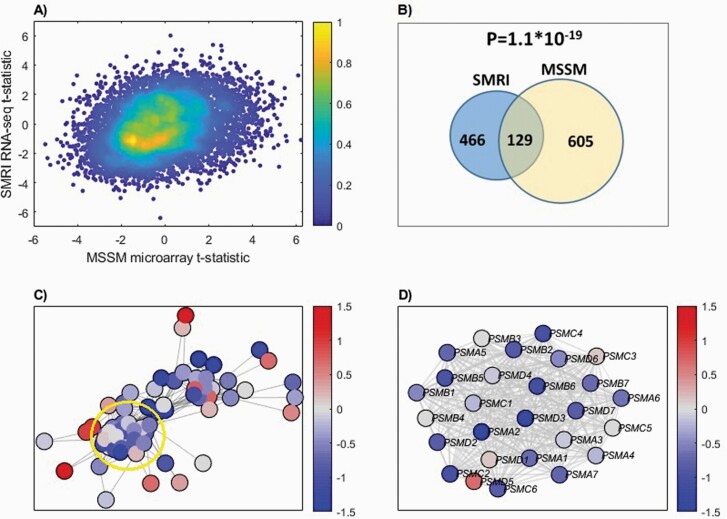
Fig. 1.
(A) Binned density scatter plot comparing the t-statistics for case versus control differential expression between the independent MSSM replication cohort assayed on microarrays and the SMRI RNA-seq data; correlation between the statistics is .28 (P = 4.7 × 10−133). The colorbar represents the density in each cell, calculated by voronoi procedure48 and normalized to values between 0 (minimal density) and 1 (maximal density). (B) Hypergeometric P-value calculation for the intersection between SMRI and MSSM down-regulated genes. The 986 SMRI and 794 MSSM down-regulated genes were intersected with the 7498 genes that are present in both cohorts, yielding 595 SMRI and 734 MSSM down-regulated genes, with 129 shared genes. (C) SMRI Differential expression network view for Ubiquitin-Proteasome Dependent Proteolysis superPathway. A node’s color corresponds to the deviation of expression from the control samples group, in terms of standard deviation units. The edges represent STRING database co-expression relations. Only genes that have co-expression relations with other genes in the network are displayed. A subgroup of highly interconnected genes, coding for proteasome subunits, is circled (D) Zoom in on proteasome subunits. The same plot as in (C), for a subgroup of highly interconnected genes coding for proteasome subunits (circled in C).