Figure 4.
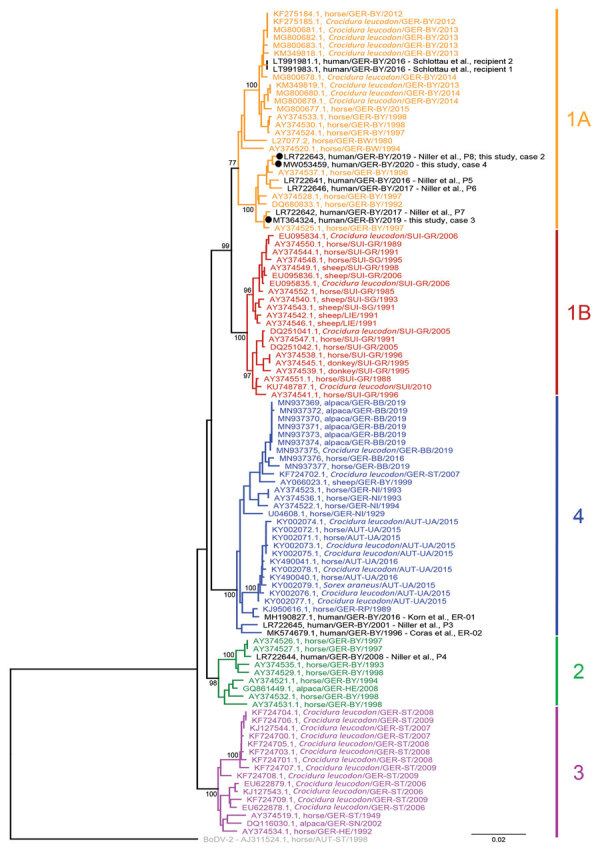
Phylogenetic analysis of BoDV-1 nucleotide sequences from virus-endemic areas, Germany. Shown are partial bornavirus sequences (nucleoprotein gene to phosphoprotein gene, 1,824 nt, representing genome positions 54–1877 of BoDV-1 reference genome U04608), including BoDV-1 sequences from animals and humans in virus-endemic regions in Germany, Austria, Switzerland, and Liechtenstein. BoDV-2 was used as an outgroup. Analysis was performed by using the neighbor-joining algorithm and the Jukes–Cantor distance model in Geneious Prime (https://www.geneious.com) and the tree was rooted for the VSBV-1 clade. Human sequences are indicated in black. Sequences of cases 1–4 included in this study are indicated in bold. Values at branches represent support in 1,000 bootstrap replicates. Only bootstrap values >70 at major branches are shown. Cluster designations, host, and geographic origin are indicated according to previously published studies (2,7,8,12,17–23). Colors and numbers at right indicate clusters. Scale bar indicates nucleotide substitutions per site. AUT, Austria: UA, Upper Austria; ST, Styria. GER, Germany: BB, Brandenburg; BW, Baden-Wuerttemberg; BY, Bavaria; HE, Hesse; NI, Lower Saxony; RP, Rhineland-Palatinate; SN, Saxony; ST Saxony-Anhalt. LIE, Liechtenstein. SUI, Switzerland: GR, Grisons; SG, St. Gall.
