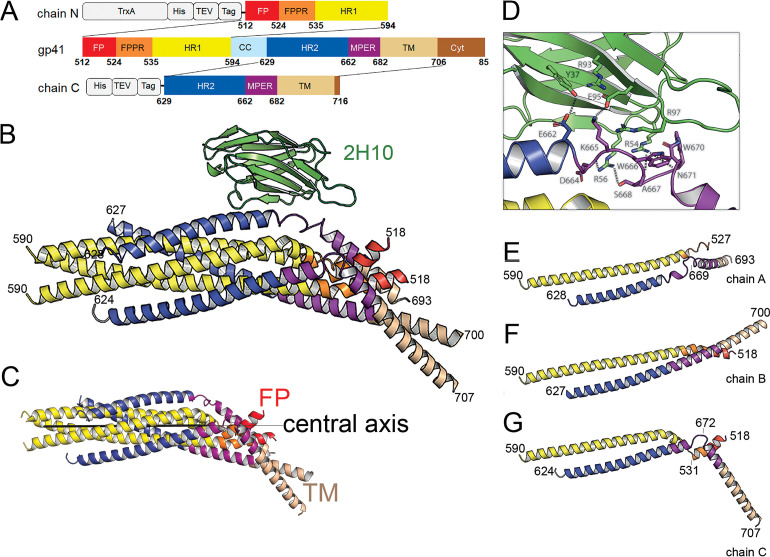
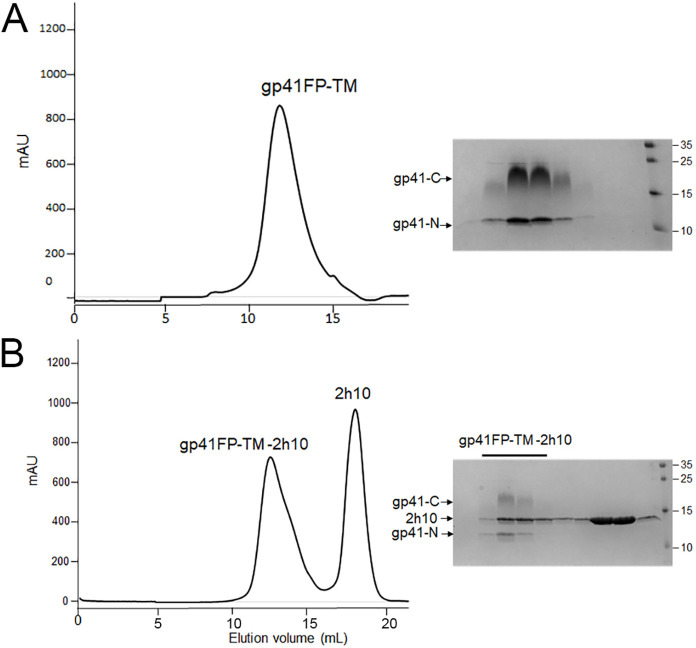
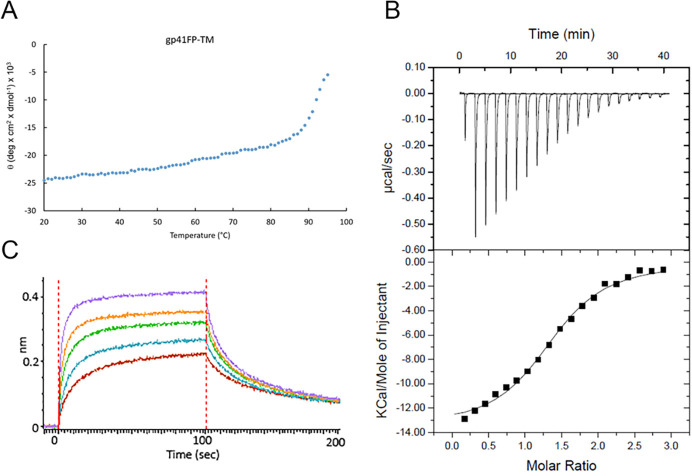
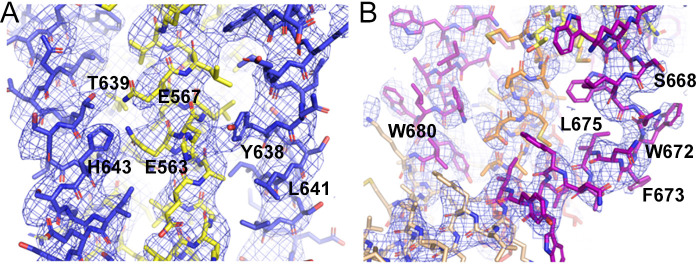
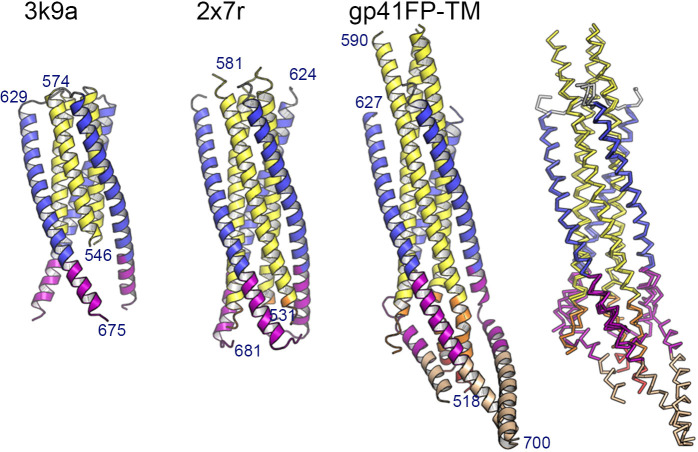
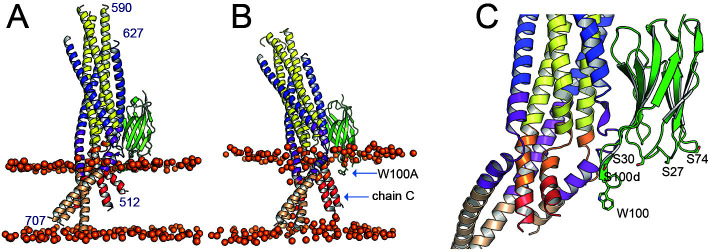
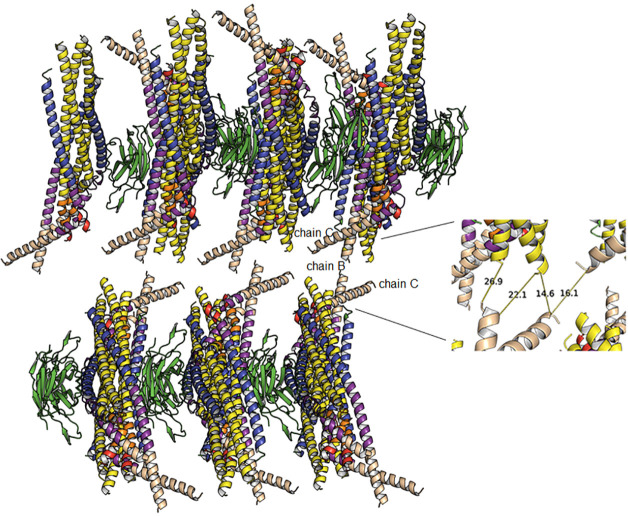
Figure 1. Crystal structure of gp41FP-TM in complex with 2H10.
(A) Schematic drawing of gp41 and expression constructs of gp41 chains N and C. Sequence numbering is based on the HIV-1-HBX2 envelope gp160 sequence. Color coding is as follows: FP, fusion peptide, red; FPPR, fusion peptide proximal region, orange; HR1, heptad repeat region 1, yellow; HR2, heptad repeat region 2, blue; MPER, membrane proximal external region, violet; TM, transmembrane region, beige; CC, cys loop region, light blue and cyt, cytoplasmic domain brown. Expression tags used are TrxA, thioredoxin fusion protein, His, His-tag, TEV, TEV protease cleavage sequence, Tag, chain N contains a Flag-tag (DYKDDDDK sequence) and chain C an N-terminal enterokinase cleavage site (DDDDK). (B) Ribbon presentation of gp41TM-FP in complex with 2H10. Color-coding of the different segments is as indicated in the gp41 scheme (A), the 2H10 nanobody is colored in green. (C) Ribbon presentation of gp41TM-FP including the core six-helical bundle trimer axis (black line) revealing the different orientations of FP and TM. (D) Close-up of the interaction of gp41FP-TM with 2H10. Residues in close enough contact to make polar interactions are shown as sticks. (E, F, G) Ribbon diagram of the individual protomers named chain A, B, and C. Residues within the FPPR and MPER hinge regions are indicated.