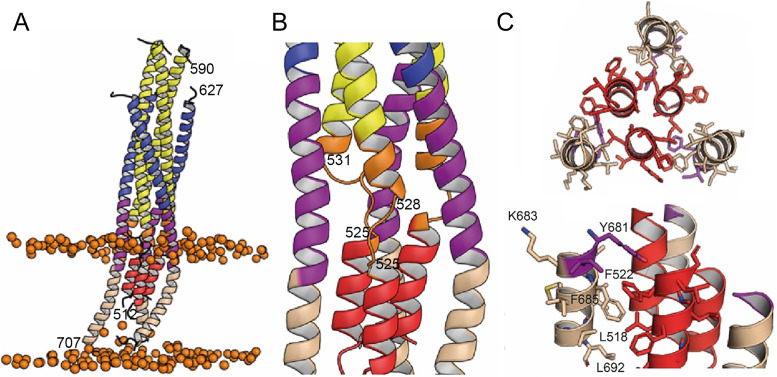
Figure 4. Interactions within the final post fusion conformation of gp41FP-TM modeled by MD.
(A) Model of gp41FP-TM (Figure 1—figure supplement 7C) after 1µs MD simulation in a bilayer. Phosphate groups of the phospholipids are shown as orange spheres to delineate the membrane boundaries. (B) Close-up on the MPER and FPPR flexible regions. (C) Close-up of the interaction of FP (residues 514–524) and TM (residues 681–692) viewed along the three-fold axis from the N-terminus indicating an intricate network of hydrophobic interactions (left panel) and from the side (right panel). Interacting side chains are labeled and shown as sticks.
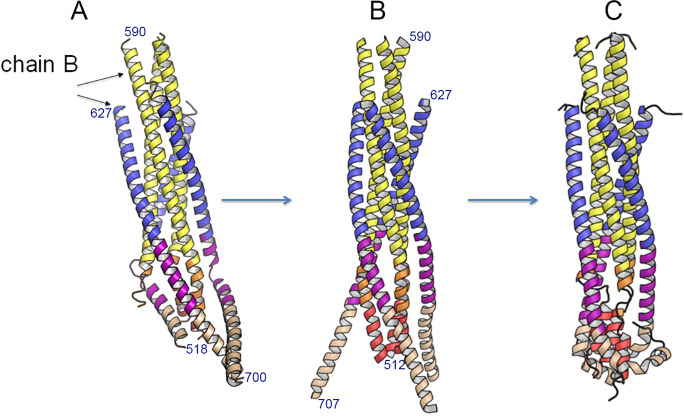
Figure 4—figure supplement 1. Modeling a post fusion conformation by MD simulation.
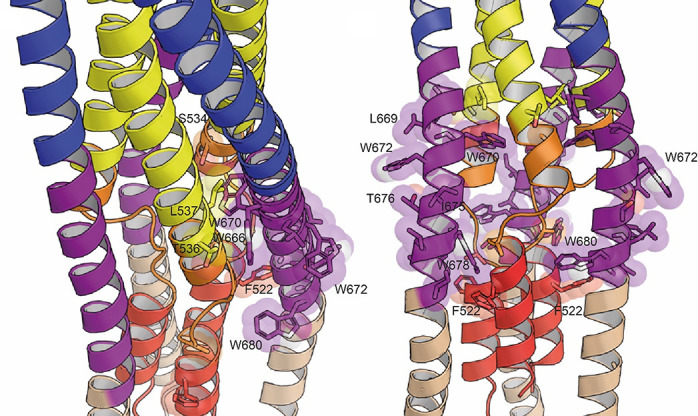
Figure 4—figure supplement 2. Positions of the conserved tryptophan residues of MPER in the post fusion model.