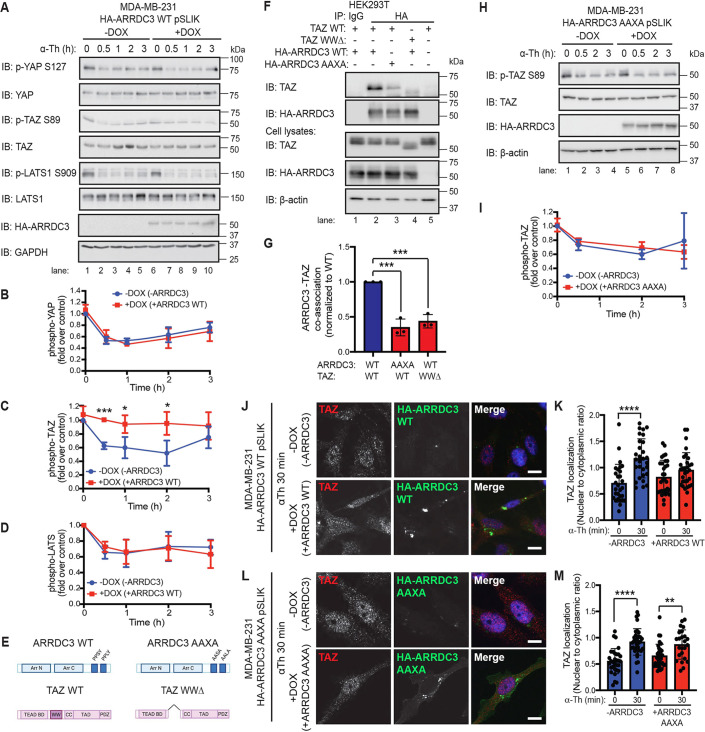
Fig. 6.
ARRDC3 re-expression blocks thrombin-mediated TAZ dephosphorylation and nuclear localization through co-association between ARRDC3 and TAZ. (A–D,H,I) MDA-MB-231 wild-type (WT: A–D) and AAXA mutant (H,I) HA–ARRDC3 pSLIK cells were treated with doxycycline (DOX) to induce ARRDC3 expression and stimulated with 10 nM α-thrombin (α-Th) for various times. Samples were immunoblotted (IB) using antibodies against the indicated proteins. Results are represented as the fold-change in YAP phosphorylation (B), TAZ phosphorylation (C,I), and LATS phosphorylation (D) relative to 0 min −DOX control. Data are mean±s.d., n=3. Statistical significance was determined by unpaired t-test at each time point. (E) Illustration of constructs of ARRDC3 and TAZ, with domains indicated (BD, binding domain; CC, coiled-coils; TAD, topologically associating domain; PDZ, PDZ domain; WW domain deletion, WWΔ). Illustration created with BioRender.com. (F,G) HEK293T cells transiently expressing wild-type and mutant constructs were immunoprecipitated (IP) with HA antibody to pulldown HA–ARRDC3. Non-specific IgG IP was used as a control. Immunoprecipitate and cell lysate input were analyzed by immunoblotting, as indicated. Results are quantified by densitometry, and co-association of ARRDC3–TAZ (G) is represented as fold over wild-type control. Statistical significance determined using one-way ANOVA with Tukey′s post hoc test. Data are mean±s.d., n=3. (J–M) TAZ subcellular localization following thrombin treatment was determined by immunofluorescence staining of endogenous TAZ (red) in MDA-MB-231 wild-type (J,K) and AAXA mutant (L,M) HA–ARRDC3 pSLIK cells; HA (green) was stained to detect ARRDC3, and DAPI (blue) for cell nuclei. Scale bars: 10 μm. (K,M) Quantification of the ratio nuclear to cytoplasmic TAZ localization. Data are mean±s.d. Statistical significance was determined by one-way ANOVA (with Tukey′s post hoc test) of each time point compared with 0 min (n=27, nine fields of view from three biological replicates). *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001.