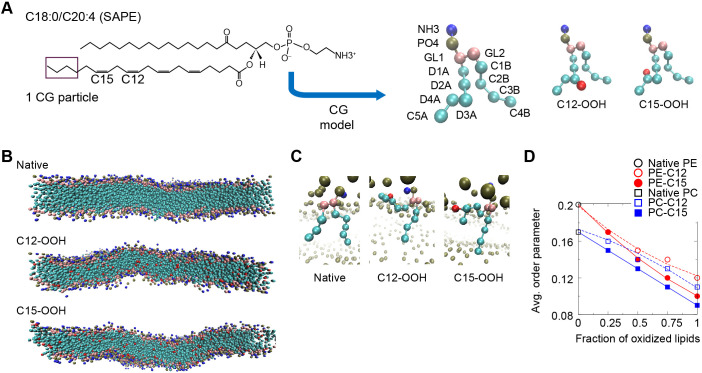
Fig. 6.
Coarse-grained molecular dynamics simulations of SAPE and SAPC lipid membranes. (A) Left panel, the chemical structure of SAPE lipid molecule with locations of the four C–C double-bonds indicated. SAPC is similar but with the NH3 group substituted by the NC3 group. Middle and right panels, coarse-grained (CG) models (4 atoms to 1 particle mapping) of unoxidized (native) SAPE lipid, oxidized SAPE with hydroperoxide functional group (-OOH) linked to a D3A particle that includes C12 (C12-OOH) and to a D4A particle that includes C15 (C15-OOH). Head-group NH3 particle in blue; PO4 particle in dark green; glycerol particles (GL1 and GL2) in pink, carbon particles in cyan (‘D’ particles contain double-bonds whereas ‘C’ particles contains only single-bonds); and OOH particle in red. (B) Equilibrated configurations of native SAPE, C12-OOH and C15-OOH bilayers after 2 μs simulation (solvent omitted for clarity). (C) Orientation of representative native and oxidized SAPE lipids in the bilayer at the end of each 2-μs simulation. (D) Mean values of order parameter computed on the basis of the angle between each tail bond and the bilayer normal, with averaging done over all the C–C bonds. The error bars were omitted as the standard errors are of the order of 0.001, or <1% of the averaged values.