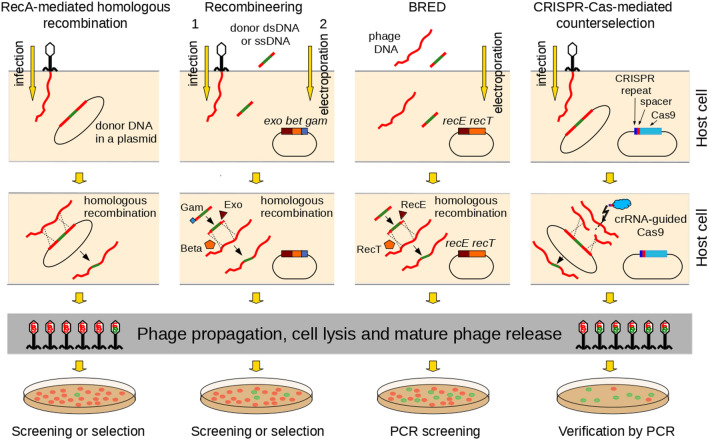
Fig. 2.
In vivo homologous recombination-based methods of bacteriophage genome modification. Proteins depicted on the scheme of recombineering represent phage λ proteins that were originally used in this method [191]. Bet and its homologs/analogs are sufficient for recombineering with ssDNA substrates as donors (reviewed in [188]). Phage proteins depicted on the scheme of BRED represent Escherichia coli prophage Rac proteins RecE and RecT, whose mycobacterial phage Che9c homologs (gp60 and gp61) were originally used in this method [198]. Recombination with the use of RecE and RecT does not require Gam (reviewed in [188]). Bet and RecT phage λ or prophage Rac single-strand annealing proteins, BRED bacteriophage recombineering of electroporated DNA, Cas CRISPR-associated proteins, Cas9 nuclease Cas9, CRISPR clustered regularly interspaced palindromic repeats, crRNA CRISPR RNA, dsDNA double-stranded DNA, Exo and RecE phage λ or prophage Rac 5′-3' exonucleases, respectively, that degrade dsDNA to expose the created 3' single-stranded ends for binding of Bet or RecT (respectively), Gam phage λ protein increasing recombination frequency by inhibiting dsDNA substrate degradation by E. coli RecBCD, PCR polymerase chain reaction, RecA bacterial recombinase RecA, ssDNA single-stranded DNA