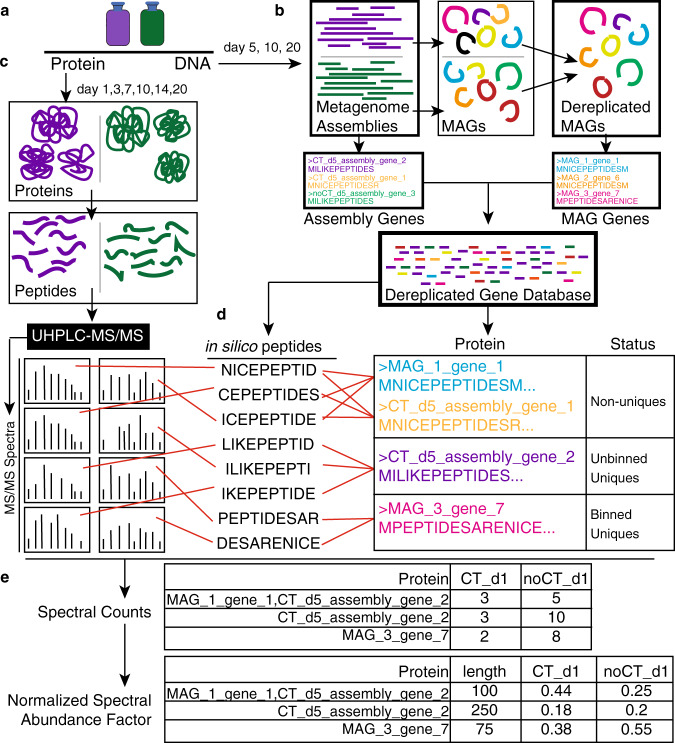
Fig. 4. Workflow for genome-resolved metaproteomics.
a DNA and proteins were sampled from triplicate live soil + CT (purple) and unamended soil (green) reactors at days 5, 10, 20 (DNA) and days 1, 3, 7, 10, 14, 20 (protein). b Metagenomes at each timepoint were obtained for both CT (purple) and unamended (green) treatments. Metagenomes were assembled and binned to obtain metagenome-assembled genomes (MAGs) across all samples. This set of MAGs was dereplicated at 99% ANI to obtain a MAG database of 155 dereplicated MAGs (Fig. 3). Using amino acid translations of genes derived from this dereplicated MAG database and remaining genes from metagenomic assemblies (on unbinned scaffolds >2500 bp), we compiled a Dereplicated Gene Database (all unique gene sequences) that served as our reference database for our metaproteomes. c Metaproteomes at each timepoint were obtained as described in Methods. d Spectral matching was carried out using obtained spectra and in silico spectra derived from the gene database. From this, proteins were classified as “non-unique” if the recruited peptides could be derived from other proteins in the database. Proteins were classified as “unbinned uniques” if they had peptides that could only be matched to the amino acid sequence derived from a metagenomic unbinned scaffold in our assembly. Proteins were identified as “binned uniques” if they had peptides that could only be matched to that amino acid sequence, and were derived from a single genome in our MAG database. e All identified proteins were quantified with label-free spectral counts. This was then corrected for protein length and sample-to-sample variation by conversion to normalized spectral abundance factor.