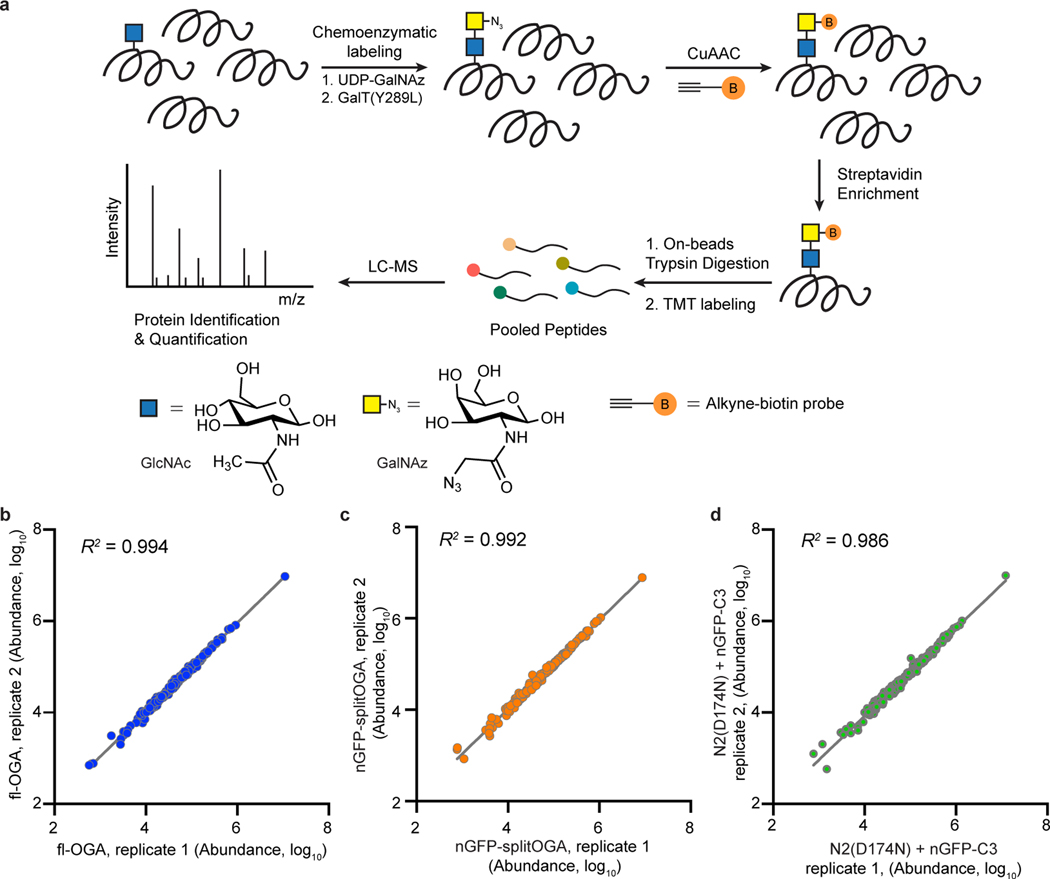
Extended Data Fig. 7. Mass spectrometry analysis on the activity and selectivity of nGFP-splitOGA on GFP-Nup62.
a, Schematic representation of the workflow of O-GlcNAcylated protein enrichment and mass spectrometry-based identification. Proteins with O-GlcNAc modification were labeled with GalNAz by GalT(Y289L)-mediated chemoenzymatic labeling, followed by a click reaction with an alkyne-biotin probe. Biotin-labeled proteome was enriched by streptavidin beads and digested by trypsin. Released peptides were labeled by TMT reagents and compiled into a single pool. Proteins were identified and quantified by LC-MS. b–d, Reproducibility of the TMT experiments of O-GlcNAcylated proteome shown in Fig. 3d,e. The signal abundances of the corresponding TMT channels for each protein were extracted and were log10 transformed for full-length OGA treatment (b, fl-OGA), nGFP-splitOGA treatment (c) and its inactive form [N2(D174N) + nGFP-C3] treatment (d) groups (n = 2 independent biological replicates).