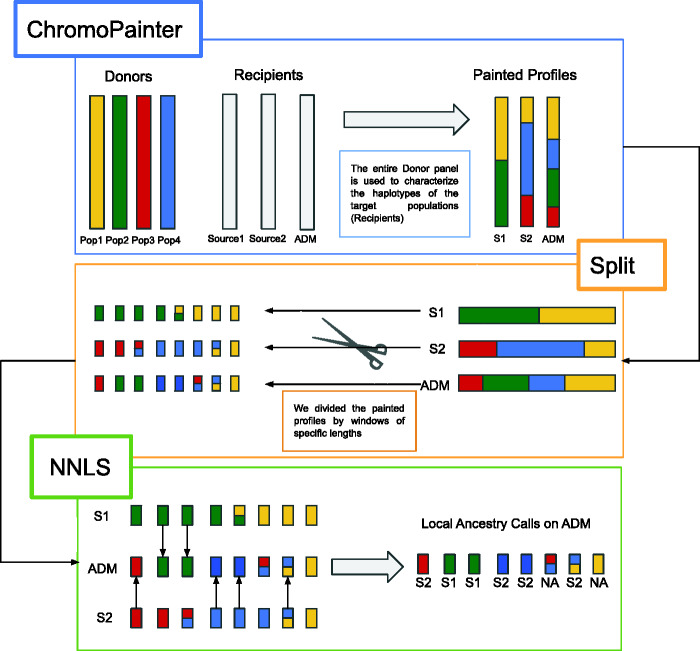
Fig. 1.
Schematic representation of WINC approach. WINC is based on the ChromoPainter/NNLS framework, with the additional step of splitting the copying vectors resulting from the ChromoPainter (CP) run before analyzing them through the NNLS step. First step: ChromoPainter run. CP identifies the closest neighbor “donor” for any “recipient” individual haplotype. ChromoPainter then reconstructs the recipient individuals as a combination of genomic segments, or chunks, “donated” by any other individual in the data set. The information is then stored in copying vectors, where, for each recipient haplotype, it is indicated which donor individual is the closest neighbour. In this way, we obtain the copying vectors of our target populations: both the sources and the admixed individuals. Second step: splitting copying vectors. We then split the copying vectors in genomic windows of the same length. Window size depends on the ancestry chunks, which in turn depends on the amount of generations since the admixture. Third step: performing NNLS analyses on the copying vector’s genomic windows obtained from the previous step. The NNLS step assigns a window to a specific ancestry, by reconstructing the painting profile of a given individual as a combination (or proportion) of copying vectors from the source individuals.