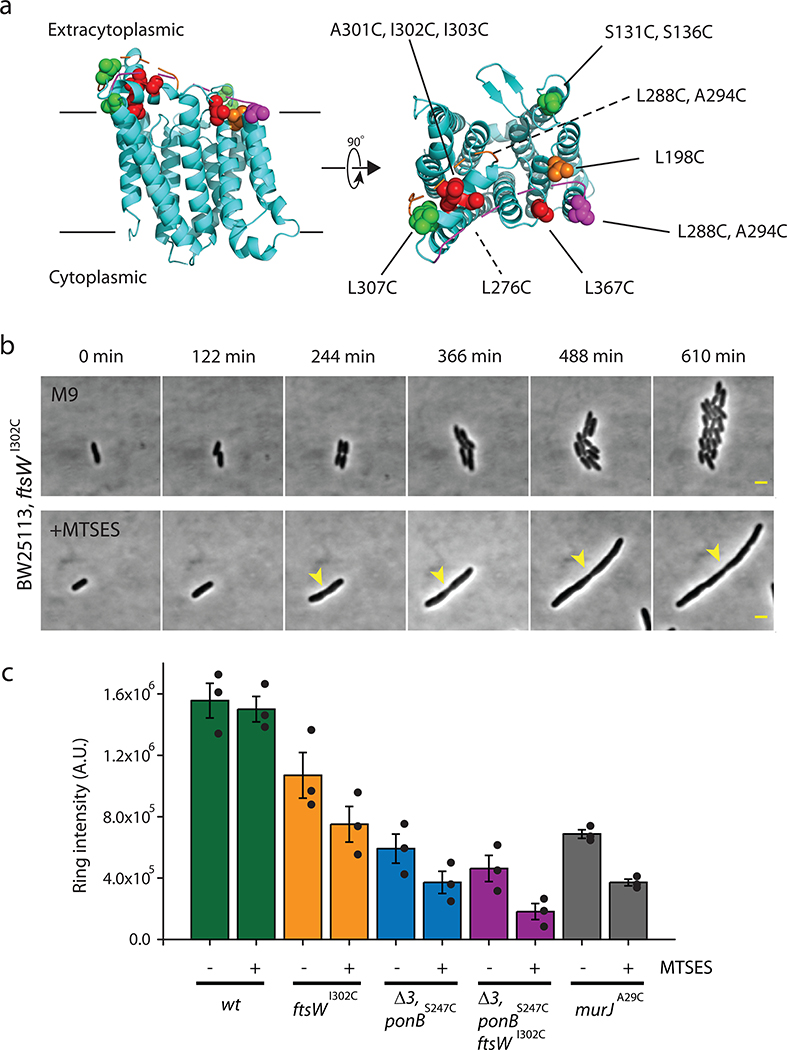
Extended Data Fig. 1. Single Cys residue substitutions in FtsW alter its function and cell morphology.
a. Homology structure of FtsW based on the structure of RodA (PDB: 6BAR)9 using Phyre 2.044. Mutated residues are labeled and color-coded according to their sensitivity to MSTES. Green: MTSES does not affect cell growth or morphology (S131C, L276C, and L307C). Orange: MTSES significantly slows down cell division and leads to elongated cells (S136C, L198C, L288C, and A294C). Red: MTSES blocks cell division completely and leads to long, chaining cells (A301C, I302C, I303C, and L367C). Magenta: Cysteine mutation causes other cell division defects such as abnormal septa and cell poles even in the absence of MTSES (L268C). b. Time-lapse images of JXY559 (ftswI302C) cells in M9 medium without (top panel) and with (bottom panel) MTSES. MTSES was added to the cell in the bottom panel 30 min prior to the first image (Supplementary Movie 1 and 2). Arrowhead marks a cell constriction that initiated early on but failed to complete. Scale bars:2 μm. c. Integrated intensity of septal NAM labeling of different strains in the absence or presence of MTSES. Error bars represent S.E.M of three independent experiments with the same imaging condition.