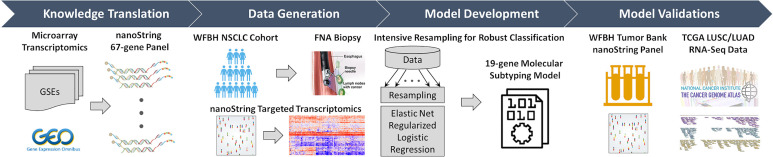
Figure 1.
Overall workflow of the C2Dx development. The development of the C2Dx NSCLC subtyping device was composed of the following four steps: 1. Knowledge translation. Four microarray-based transcriptomics datasets of NSCLC cases were extracted from Gene Expression Omnibus (GEO) and used to identify 67 subtyping-related genes, including 23 LUAD genes, 40 LUSC genes, and 4 housekeeping genes. A corresponding nanoString gene panel was established. 2. Data generation. Targeted transcriptomics data were generated using the nanoString 67-gene panel for 83 FNA biopsy samples collected from WFBH NSCLC patients, including 47 LUAD, 25 LUSC, and 11 NOS cases. 3. Model development. Intensive resampling strategy was used in elastic-net regularized logistic regression to develop the final 19-gene molecular subtyping model. 4. Model validation. The developed subtyping model was validated using the transcriptomics data of the 19 genes collected from frozen tissue from WFBH’s tumor bank using nanoString platform and from the TCGA’s RNA-Seq data generated from bulk tissues.