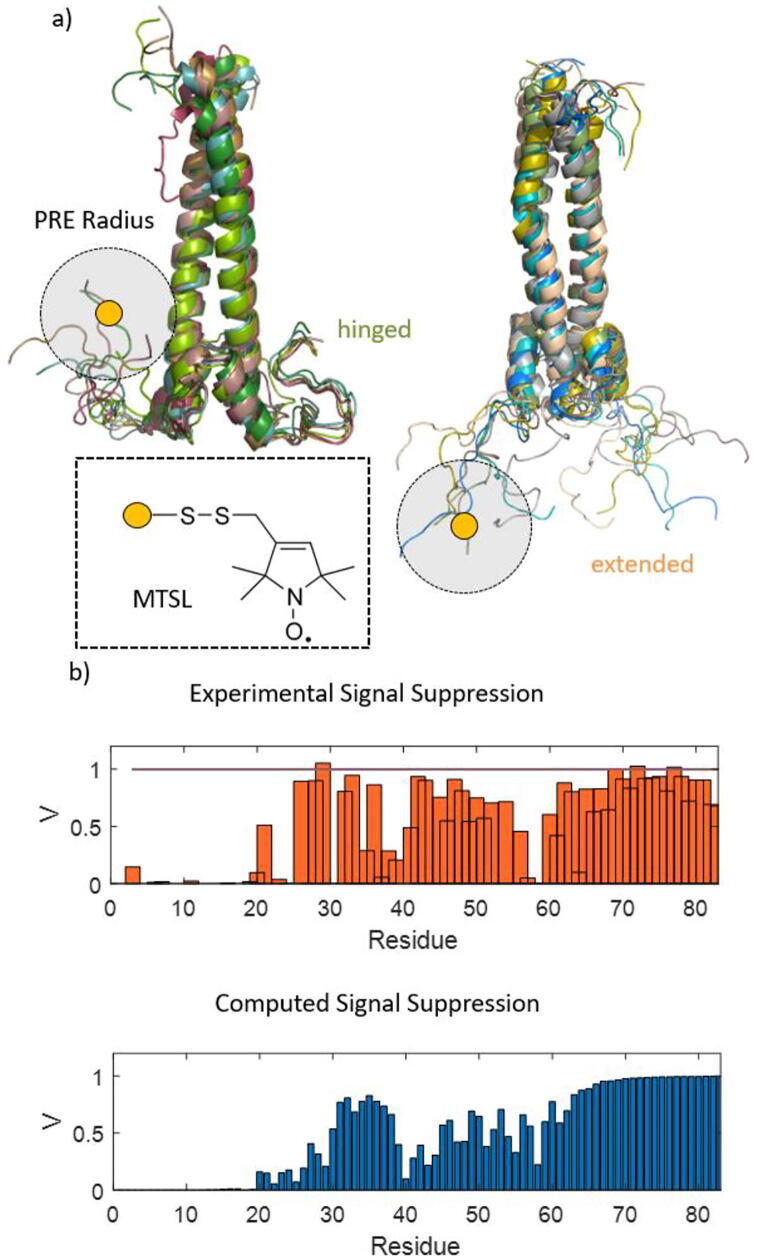
Fig. 1.
Comparison of experimental and computed PRE effects for the MAX transcription factor. a) Conformations sampled in MD simulations of the MAX:MAX homodimer. The DNA-binding domain (bottom) is intrinsically disordered in the absence of a ligand and samples a heterogeneous conformational space that includes hinged and extended conformations. If a spin-label is attached to the DNA-binding site (e.g. at the site of the yellow dot) NMR signals of amino acids in its vicinity are suppressed by PREs (effect range indicated by the grey shade). Thus, the hinged conformation would lead to the suppression of signals assigned to the remote HLH domain, while the extended conformations would not. The structure of the attached SL is indicated in the dashed box. b) Experimentally observed PRE effects as a function of residue position in MAX:MAX (top) compared to PRE effects extracted from MD simulations (bottom). The match between both data sets is good, such that the conformational ensemble sampled in the MD simulations could be verified by the experimental observations (adapted from reference [61] with permission of the publisher.) (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)