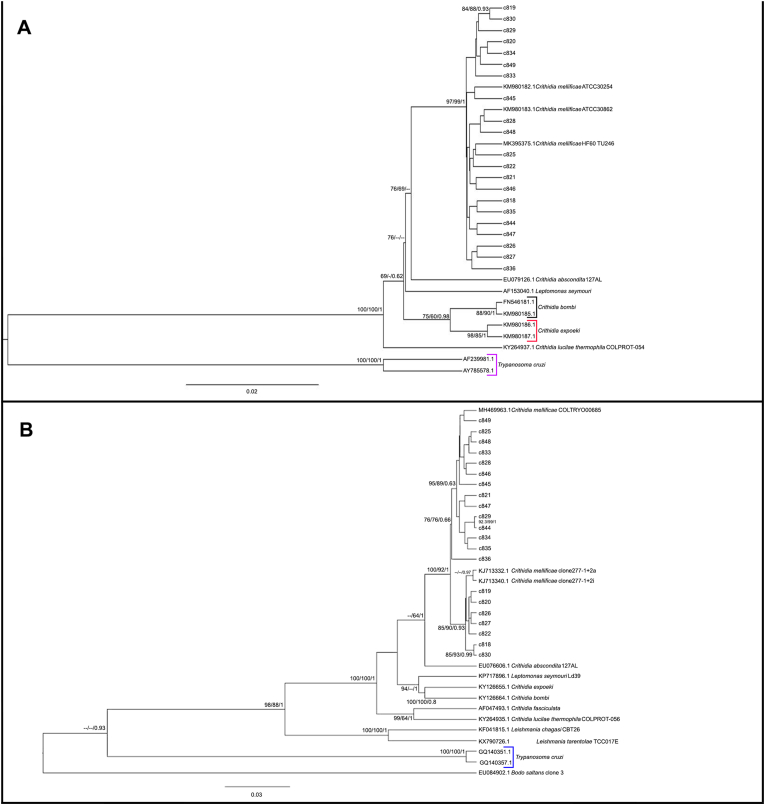
Fig. 3.
Phylogenetic analysis of V7V8 SSU rDNA and gGAPDH sequences from bat, primate, and carnivore haemocultures by maximum likelihood (ML) and Bayesian (BI) inference analyses. (A) For V7V8 SSU rDNA, the analysis was inferred using the transition-parameter model with invariant sites (TIM3 + I). (B) For gGAPDH, the analysis was inferred using the General Time Reversible with invariant plus gamma distributed sites (GTR + I + G). Maximum likelihood ultrafast and SH-aLRT bootstrap values and Bayesian posterior probabilities are shown near the nodes. The numbers at the nodes indicate support by 5000 bootstrap pseudoreplicates in the ML analysis. The scale bar shows the number of nucleotide substitutions per site. The dashes at the nodes represent bootstrap or posterior probability lower than 60 or 0.6.