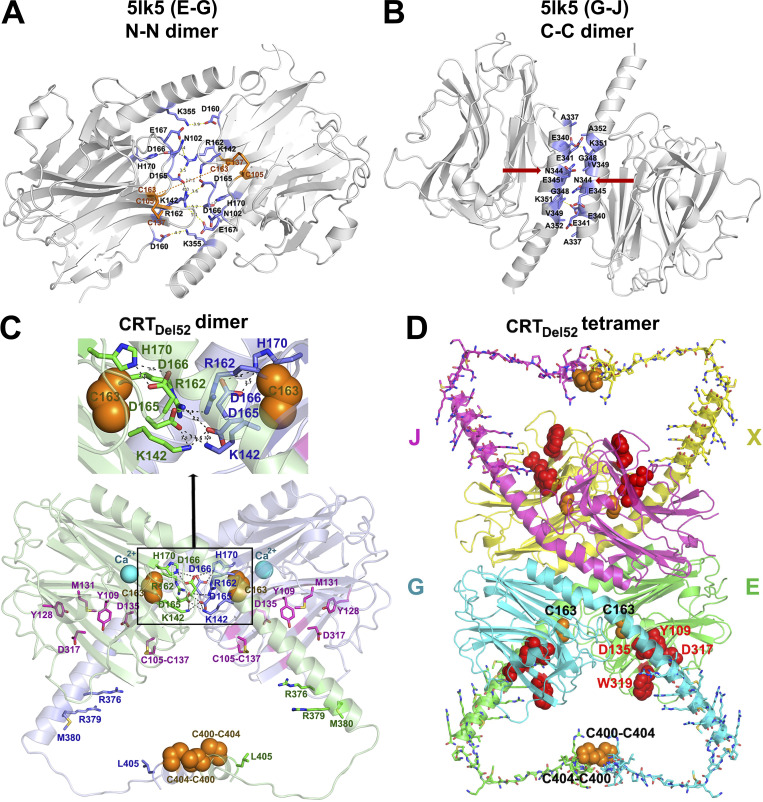
Figure 5.
Proposed structural models for CRTDel52 dimers and tetramers. (A and B) Two major dimerization modes were observed in the crystal structure of the 10-mer complex of CRT D71K mutant (Protein Data Bank accession no. 5LK5; Moreau et al., 2016): dimerization via N-domain loops rich on charged residues that form intermolecular ionic bridges (N-N dimer between subunits E-G and J-X; A) and tight packing of antiparallel α-helices (C-C dimer between subunits G-J and E-X; B). Contacting residues at C-domain helix–helix and N-domain loop–loop interfaces are shown by sticks colored blue for C atoms. Red arrows indicate N344, a glycosylation site. (C) A molecular model of a proposed CRTDel52 dimer. Subunit 1 is colored light green and subunit 2, light blue. Each subunit contains a globular N-domain and a C-domain composed of long and short α-helices connected by a 14-residue loop. The P-domain is omitted. Ca2+ ions bound to the high-affinity site are shown by cyan spheres. Residues from the carbohydrate recognition site are shown by purple sticks (for C atoms). Cysteines participating in the formation of predicted intermolecular disulfide bonds (two C400-C404) and two C163 residues are shown by orange spheres. Residues forming H-bonds and ion pairs (K142, R162, D165, D166, and H170) are shown by sticks colored green (for C atoms) for subunit E and blue for subunit G. Inset highlights interactions at the contact interface. (D) A homology model of CRTDel52 tetramer lacking P-domains. Mutated residues (367–406) from the novel C-tail of CRTDel52 are shown by sticks. Cysteine residues from each subunit predicted to form intermolecular disulfide bonds are shown by orange spheres. Residues (Y109, D135, D317, and W319) located in glycan-binding pockets are shown by red spheres.