Abstract
Phospholysine phosphohistidine inorganic pyrophosphate phosphatase (LHPP) is a new-found tumor suppressor in a variety of tumors. While, it is still unknown about its role in glioma. In this study, we found that LHPP is abnormally decreasing or absent in glioblastoma, and the low expression of LHPP is associated with poor median survival in glioma patients. Functional assay revealed that LHPP-overexpression significantly inhibited U87MG and U118MG growth in vitro and in vivo. As to the mechanism, mass-spectrometric analysis indicated that the LHPP interacting proteins were mainly enriched in regulation of energy metabolism, including Carbon metabolism, Oxidative phosphorylation, and Glycolysis. Seahorse assay and metabolites detection confirmed that LHPP-overexpression obviously impeded glycolysis and respiration in U87MG and U118MG cells. For the further study, western blot assay showed that the protein level of PKM2 at dimeric, tetrameric, and total protein, were all decreased significantly, and its enzymatic activity was decreased as well. ChIP and RNAseq integrated analysis indicated that the decreased protein level of PKM2 was independent of PKM2 transcription, and LHPP did not reprogram transcription level of metabolic genome. Co-IP and immunofluorescence assay manifested that LHPP interacted with PKM2, and this interaction interfered the protein stability, then induced ubiquitin-mediated degradation of PKM2. Rescue assay confirmed that restoring the expression of PKM2 effectively reversed the restrained energy metabolism and the inhibited cancer cell growth caused by LHPP-overexpression in U87MG and U118MG cells. Taking together, we demonstrated that LHPP impedes the glycolysis and respiration during energy metabolic process via inducing ubiquitin-mediated degradation of PKM2, thus inhibits the growth of glioblastoma.
Keywords: LHPP, glioblastoma, energy metabolism, PKM2, ubiquitination degradation
Introduction
Glioma is the most common primary brain tumor in human central nervous system, among which glioblastoma multiforme (GBM) is the most malignant pathological type [1,2]. At present, the therapeutic methods for GBM include maximal safe surgical resection, followed by high-dose radiation and standard chemotherapy [3]. Unfortunately, due to the complicated and hierarchical internal cellular organization of tumor entities, GBM often exhibits refractory to conventional therapies, and caused an extremely poor clinical prognosis [4-6]. Although there were some newly and advanced immunotherapy and molecularly targeted approaches, we still unable to effectively prolong the overall survival of GBM patients [7,8]. Discovering new biomarkers is beneficial for improving the diagnostic accuracy and individualized treatments, which encourages us to find much more novel glioma-specific or glioma-related biomarkers [9].
Phospholysine phosphohistidine inorganic pyrophosphate phosphatase (LHPP), is initially discovered in swine brain tissue [10], and it is evolutionarily conserved from worm to human [11]. LHPP is a multifunctional gene which impact on the structural of gray matter, regional brain activity, and pathophysiological progression in major depressive disorder [12-14]. Sravanth KH firstly reported that LHPP is a tumor suppressor in hepatocellular carcinoma [15]. Subsequently, studies indicated that LHPP also exhibited inhibitory effect on papillary thyroid cancer, bladder cancer, melanoma, cervical cancer, and colorectal cancer [16-20]. While, it is still unclear the role of LHPP in glioma.
In this study, we found that LHPP is abnormally decreased or not expressed in glioblastoma, and the lower LHPP level predicted a shorter median survival in glioma patients. LHPP-overexpression in glioblastoma cells exhibits an obviously inhibitory effect on cancer growth in vitro and in vivo. The mechanism studies revealed that LHPP interfered the protein stability of PKM2, and then induced ubiquitin-mediated degradation of PKM2, which further impedes glycolysis and respiration during energy metabolic process in glioblastoma cells. Meanwhile, restoring the expression of PKM2 effectively reversed restrained energy metabolism and the inhibited cancer cell growth caused by LHPP-overexpression in glioblastoma cells. This study provides scientific clues for the biological role and molecular mechanism of LHPP in glioblastoma, which may help with the future diagnosis and treatment of glioblastoma.
Materials and methods
Databases
The Human Protein Atlas (https://www.proteinatlas.org/) was used to analyze expression profile of LHPP in human tissue and cancer cell lines. Transcriptomics datasets, TCGA-GBM and TCGA-GBMLGG, in The Cancer Genome Atlas (TCGA, https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga), and mRNAseq datasets, batch 1 and batch 2, in Chinese Glioma Genome Atlas (CGGA, http://www.cgga.org.cn/) were downloaded and used for evaluating the clinical relevance of LHPP expression in glioma.
Cells and cell culture
Human GBM cell lines, U87MG, U251MG, T98G, and LN229 were purchased from the Chinese Academy of Sciences cell bank (Shanghai, China), U118MG and A172 were purchased from Obio Technology Co., LTD (Shanghai, China). The normal human astrocyte (HA, ScienCell) was donated by Tao-Liang Chen (Zhujiang Hospital, Southern Medical University). GBM cell lines were culture in DMEM/F12 (12100-046, Gibco, USA) which containing 10% Fetal Bovine Serum (10099141C, Gibco, USA), HAs were cultured in astrocyte medium (#1801, ScienCell, USA) at 37°C in a humidified incubator with 5% CO2.
Glioma tissues microarray and immunohistochemistry analysis (IHC)
A glioma tissue microarray slide (Wuhan Servicebio Technology CO., LTD, Wuhan, China), which containing 94 cases was used for LHPP immunohistochemistry (IHC) staining. The slide was dried at 60°C for 1 hour. Then, the slide was deparaffinized with xylene, and rehydrated with gradient alcohol. Afterwards, Tris-EDTA buffer was used to conduct antigen retrieval, and 3% H2O2 solution was used to eliminate endogenous peroxidase activity. After that, the slide was then incubated with LHPP antibody (1:400, PA5-52658, Invitrogen, USA) at 4°C overnight. After that, the antibody was detected with Polink-2 plus polymer HRP detection system (ZSGB Bio, China). Images were taken under Pannoramic 250 (3D HISTECH, Hungary). The staining score of LHPP was evaluated with Image-Pro Plus software (Media Cybernetics, USA). Cases which complete negative staining were defined as LHPP score 0, cases which containing less than 10% positive cells (rare) were defined as LHPP score 1, cases which containing 10%~70% positive cells (not rare) were defined as LHPP score 2, cases which containing more than 70% positive cells (strong) were defined as LHPP score 3. Cases with score 0 and 1 were defined as LHPP IHC low score, cases with score 2 and 3 were defined as LHPP IHC high score.
Plasmids, lentivirus and transfection process
The LHPP plasmids, PKM2 plasmids, empty vector plasmids, lentiviral particles with LHPP, lentiviral particles with PKM2, and lentiviral particles with empty vector were all purchased from IGeneBio (Guangzhou, China). Lipofectamine 3000 Transfection Reagent (L3000008, Thermo Fisher, USA) was used for plasmids transfection according to the manufacture’s protocol. For lentiviral transfection, cells were incubated with lentivirus particles for 8-12 hours in serum-free medium, and then replaced with new medium. Puromycin (Solarbio, China) was added at a final concentration of 2 ug/mL for selecting the stable expression cells. The overexpression efficiency was confirmed by Western blotting.
Cell proliferation test
CCK-8 assay, EdU assay were performed for studying the effect of LHPP on cell proliferation in vitro. For CCK-8 assay, Enhanced Cell Counting Kit (CCK)-8 (C0041, Beyotime, China) was used. 2000 cells were seeded in 96-well plate per well for 8~12 hours. At the subsequent five days, OD450 was measured after cells were incubated with CCK-8 reagent for 1 hour at 37°C. For EdU assay, BeyoClick EdU Cell Proliferation Kit with Alexa Fluor 555 (C0075L, Beyotime, China) was used. Cells were culture in 12-well plate overnight, then treated with 20 μM EdU solution for two hours at 37°C. 4% PFA solution was used to fix cells for 10 minutes. Then, 0.5% Triton X-100 solution was used to permeabilize cells for 15 minutes. Following, wash the cells with PBS for thrice. After that, cells were incubated with Click Reaction Solution avoiding light for 30 minutes at room temperature. Finally, cells were rinsed, and stained with DAPI. The results were analyzed under Leica DMI3000B fluorescence microscope (Leica, Germany).
Cloning formation assay
For cell clone formation assay, 2000 cells were seeded in 6-cm dish for 12 hours. Then, lentiviral particles with LHPP, LHPP plus PKM2, or empty Vector were added for transfection process. After that, cells were allowed to growth until 14 days. Then, remove the medium and the cells were stained with crystal violet solution for 30 mins at room temperature. After that, remove the solution and wash the cells with PBS softly, remove the PBS solution and the cell clone were taken picture.
Intracranial glioma xenograft model
U87MG-Luc cells were transfected with lentiviral particles with LHPP, LHPP plus PKM2, or lentiviral particles with empty vector, to construct U87MG-Luc LHPP-overexpression cells, U87MG-Luc LHPP plus PKM2-overexpression cells, and corresponding control cells. 3×106 cells were stereotactically injected into the 4~6 weeks old male BALB/cA nude mice (HFK BIOSCIENCE, China). Then, the cells were allowed to grow in vivo for 14 days. After that, the status of tumor growth in vivo was measured with an IVIS (IVIS Lumina II, USA). 3 mice each group were used to take the hole brain tissue for paraffin sections, and the rest 6 tumor-burdened mice each group were used for survival assay.
Mass-spectrometric (MS) analysis for LHPP complexes
LHPP-overexpression U87MG and U118MG cells were used to perform mass-spectrometric analysis. Anti-Flag antibody was used to precipitate Flag-LHPP-interacting proteins complex, and used for SDS-PAGE procedure. Then, the protein bands were excised from SDS-PAGE gel, Milli-Q water was used to rinse the bands, and then destained bands with 25 mM NH4HCO3 and 50% acetonitrile. After that, proteins were following reduce, alkylate, and digest by 10 mM dithiothreitol, 55 mM iodoacetamide, and 10 ng/ul trypsin, respectively. Then, the digested peptides were desalted via ReproSil-Pur C18-AQ column (Thermo Scientific, USA). 2% acetonitrile (in 0.1% formic acid solution) and 80% acetonitrile (in 0.1% formic acid solution) were used for a gradient process. MS analysis was conducted with Q Exactive plus mass spectrometer (Thermo Scientific, USA).
Identification of LHPP-interacting proteins and bioinformation analysis
The raw data from MS analysis were converted to MGF data by Proteome Discoverer. Peptide masses identification was performed by Mascot software (Matrix Science, USA) based on SwissProt human database. The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways analysis, and Protein-Protein Interactions (PPIs) was analyzed using the coincident Flag-LHPP interacting proteins in LHPP-overexpression U87MG and U118MG cells.
Glucose uptake assay
Glucose Uptake Assay Kit (#KA4086, Abnova, Taiwan) was used for the measurement of glucose uptake in U87MG and U118MG cells. 36 hours after plasmids transfected, 50000 cells/well/100 μL/96-well were plated in growth medium for 4-6 hours before experiments. Then, treat the cells as desired, and add 10 μL/well 2-DG and incubate at 37°C for 30 minutes. Wash and lyse cells, 50 μL/well 2-DG Uptake Assay working solution was added in, then incubate cells at RT for 90 minutes. After that, the OD ratio at 570/610 nm was measured under spectrophotometer (1510, Thermo Fisher, USA).
Pyruvate assay, lactate assay, and ATP assay
Pyruvate Assay Kit (BC2205, Solarbio, China), Lactate Assay Kit (BC2235, Solarbio, China), and ATP Assay Kit (BC0300, Solarbio, China) were used to measure the concentration of pyruvate, lactate, and ATP level, respectively. After plasmids were transfected, cells were cultured for 48 hours, and then used for subsequent procedure according to the product manual. Relative absorbance was measured under spectrophotometer (1510, Thermo Fisher, USA). The content of pyruvate, lactate, and ATP were calculated according to the product manual.
Oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) assay
Seahorse XF96 Extracellular Flux Analyzer was used for measuring cell oxygen consumption rate (OCR) and extracellular acidification rate (ECAR). After plasmids were transfected, cells were cultured for another 24 h. Then, cells were plated in XF96 cell culture plates (Seahorse Bioscience, USA), and incubated for 8 h at normal cell incubator. After that, cells were equilibrated with bicarbonate-free buffered DMEM for 1 h without CO2 immediately before XF assay. The glycolytic rate assay was performed in XF Base Media without phenol red containing 5 mM HEPES, 10 mM glucose, 1 mM sodium pyruvate, and 2 mM L-glutamine, and the following inhibitors were added at the final concentrations: rotenone/antimycin A (0.5 μM each) and 2-deoxy-glycose (50 mM). The mitochondrial stress test was performed in XF Base Media containing 10 mM glucose, 1 mM sodium pyruvate, and 2 mM L-glutamine, and the following inhibitors were added at the final concentrations: oligomycin (1 μM), carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone (1 μM), and rotenone/antimycin A (0.5 μM each).
PKM2 enzymatic activity assay
Pyruvate Kinase Assay Kit (BC0540, Solarbio, China) was used to determine PKM2 enzymatic activity. After plasmids were transfected, cells were cultured for 48 hours. Then, cells were collected into the centrifuge tube and the supernatant was discarded after centrifugation. Then, 5×106 cells were added with 1 mL extracting solution and the cells were destroyed by ultrasound (ice bath, 200 W, ultrasound 3 s per time, 10 s interval, repeat 30 times). After centrifugation at 8000 g at 4°C for 10 min, the supernatant was taken and placed on ice for measurement. Working solution was real-time prepared according to manufactures’ protocol. An aliquot of 10 μL cell lysate was used for the assay. Absorbance at 340 nm on a microplate reader was recorded at 0 min (A1) and at 2 min 20 s (A2). PK (U/104 cell)=5.226×(A1-A2).
Integrated RNA sequencing (RNAseq) and ChIP sequencing (ChIPseq) analysis
For RNAseq, U87MG cells were transfected with LHPP plasmids or empty vector, and then cultured for 48 hours. After that, the total RNA was extracted, and measured with quality by Bioanalyzer (Agilent, USA) and RNA-seq libraries. Then, RNA samples were enriched for stranded poly (A) mRNA, and sequenced on IIIumina HiSeq Novaseq 6000. The raw trimmed reads were aligned to the reference genome (GRCh38).
For ChIPseq, U87MG cells were transfected with LHPP plasmids, and cultured for 48 hours. Then, fix the cells with 1% formaldehyde solution, and sonicated the cells for obtaining soluble chromatin. Flag-LHPP-DNA complexes were separated from 20 μg chromatin by adding 5 μg anti-Flag antibody (#14793, CST, USA). ChIP-seq reads were trimmed and aligned to the hg38 genome by trim-galore and bwa. Flag-LHPP binding regions were identified by using MACS2. Peaks overlapping a 3 kb window around a ENSEMBL v99 transcription start site (2.5 kb upstream, 0.5 kb downstream) were tagged as promotors. Flag-LHPP binding sites were determined using bedtools. Alignment data of Flag ChIPseq and RNAseq were performed to check the consensus between Flag-LHPP bound peaks and target gene transcription status.
Quantitative real-time PCR (qRT-PCR) assay
PKM2 mRNA level were measured by Quantitative-PCR assay. After plasmids were transfected, cells were cultured for 48 hours. Then, total RNA was extracted by TRIzol Plus RNA Purification Kit (12183555, Invitrogen, USA) according to the instructions. PrimeScript RT Reagent Kit (RR037B, TaKaRa, Japan) was used to construct cDNA. qRT-PCR analysis was performed by Platinum SYBR Green qPCR SuperMix-UDG w/ROX kit (11744100, Thermo, USA) according to the manufacturer’s protocol. Quantitative PCR reactions were repeated at least three times. The expression level was normalized to GAPDH. The primer sequence information used in this part as follows: PKM2: forwards primer is 5’-ATCGTCCTCACCAAGTCTGG-3’, and the reverse primer is 5’-GAAGATCCACGGTACAGGT-3’; GAPDH: forwards primer is 5’-CTCATGACCACAGTCCATGCC-3’, and the reverse primer is 5’-TTCCCGTTCAGCTCAGGGAT-3’.
Formaldehyde cross-linking [21]
Formaldehyde Cross-linking was performed for studying the PKM2 dynamic populations. After plasmids were transfected, cells were cultured for 48 hours. Then, cells were incubated with 0.4% paraformaldehyde (PFA) solution for 30 mins at 37°C. After that, 1.25 M glycine was added (final concentration of 125 mM) to terminate the cross-linking reaction. Cells were washed with PBS, and resuspended in lysis buffer (50 mM Tris, 150 mM NaCl, 10% glycerol, 1% NP-40, and 5 mM EDTA) of which containing protease inhibitor (P1011, Beyotime, China). Finally, cells lysates were centrifuged at 15000 rpm for 15 mins, and the supernatants were collected for western blot analysis.
Western blot
Total protein in cells was extracted with RIPA Lysis Buffer (P0013B, Beyotime, China) of which containing protease inhibitor (P1011, Beyotime, China). Protein concentration was measured with BCA protein assay kit (PC0020, Solarbio, China). Proteins were separated by SDS-PAGE electrophoresis, and transferring to PVDF membrane. After antigen blocking with 5% BSA solution (ST023, Beyotime, China), the membranes were incubated with primary antibodies overnight at 4°C. Then, membranes were rinsed thrice with cooled TBST solution for three times, then incubate the membranes with HRP-conjugated secondary antibody at room temperature for 1 h. Membranes were washed cooled TBST solution for three time, and then reacted with Immobilon Western Chemiluminescent HRP Substrate (P90719, Millipore, USA) for 10-15 s. Then the proteins in membranes were detected with chemiluminescence imaging system (Bio-Rad, USA).
The antibodies used in this part: LHPP (sc-376648, Santa Cruz, USA; PA5-52658, Invitrogen, USA), PKM2 (#4053, CST, USA), GAPDH (ab8245, Abcam, USA), beta Tubulin (ab6046, Abcam, USA), beta Actin (ab8226, Abcam, USA).
Co-immunoprecipitation (Co-IP)
Cells were collected and lysed using RIPA Lysis Buffer (P0013B, Beyotime, China) of which containing protease inhibitor (P1011, Beyotime, China). 2 ug antibody was added into the lysate and incubated overnight at 4°C, then, 20 μL Protein A/G PLUS-Agarose beads (sc-2003, Santa Cruz, USA) was added into the lysate and rotated for 2~4 hours at 4°C. The beads-antibody-protein complexes were washed with pre-cooled PBS solution for three times, and then boiled for subsequent western blot analysis.
The antibodies used in this part: LHPP (sc-376648, Santa Cruz, USA), PKM2 (#4053, CST, USA), Flag (#8146 and #14793, CST, USA), Ubiquitin (#3936, CST, USA).
Immunofluorescence (IF) staining
U87MG, U118MG, LHPP-overexpression U87MG, LHPP-overexpression U118MG, and HA cells were used for immunofluorescence staining. Cells were cultured in Millicell EZ slide (R0EB54201, Millipore, USA), and fixed with 4% Paraformaldehyde (PFA) solution for 15 mins at room temperature. Then, wash the cells with PBS thrice, and 0.3% Triton X-100 solution was used for permeabilization for 15 mins at room temperature. After that, 10% BSA (ST025, Beyotime, China) was used for antigen blocking for 2 hours at room temperature. Following, cells were incubated with primary antibodies for 16 hours at 4°C. Then, cells were rinsed by PBS solution thrice, and then incubated with secondary antibodies at room temperature for 2 hours. Lastly, cells were rinsed by PBS solution thrice again, and mounting medium with DAPI was added for nuclear staining. Images were taken under fluorescence microscope (DM 3000B DFC425C, Leica, Germany.
The antibodies and agent used in this part: LHPP (sc-376648, Santa Cruz, USA), Flag (#8146, CST, USA), PKM2 (#4053, CST, USA), Goat polyclonal Secondary Antibody to Mouse/Rabbit lgG-H&L (488/555/647, Abcam, USA). Mounting Medium with DAPI (ab104139, Abcam, USA).
Statistical analysis
GraphPad Prism 8 (GraphPad Software Inc., USA) was used to perform the statistical analysis and statistical chart making. Quantitative data was analyzed by unpaired t test, the survival rate and prognosis were estimated using Kaplan-Meier method. Each group of experiments was repeated at least three times, SME was calculated to indicate the variation within each experiment and data, and the values represent mean ± SME. P<0.05 is considered to be statistically significant.
Results
Abnormal decreased expression of LHPP in human glioblastoma, and is correlated with a poor prognosis
We accessed The Human Protein Atlas database, the LHPP protein expression overview and mRNA consensus expression overview both indicate that LHPP is high expression in normal brain tissue, and the mRNA expression level of LHPP is relatively low in glioma cell lines (U251MG, U87MG, and U138MG) among a total of 64 cancer cell lines (Figure S1A-C). These results suggested LHPP is highly expressed in normal brain tissue, and low expressed in glioma. This abnormal decrease expression level of LHPP may be related to the incidence of glioma.
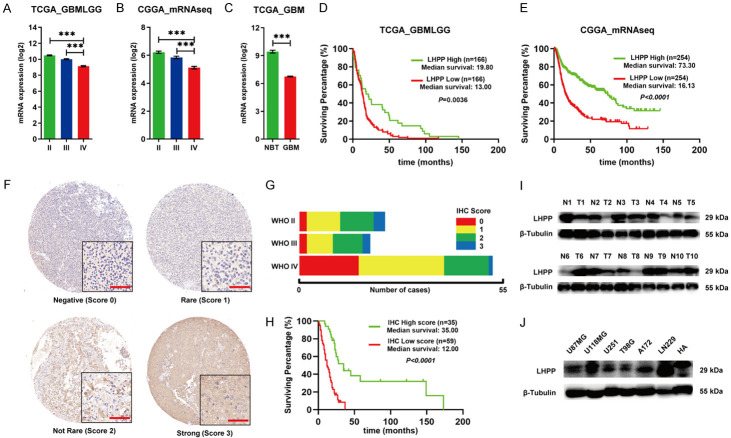
Then, we explored the expression profile of LHPP in glioma based on two open databases, The Cancer Genome Atlas database (TCGA) and Chinese Glioma Genome Atlas (CGGA). TCGA_GBMLGG and CGGA_mRNAseq database show that mRNA level of LHPP was significantly lower in WHO grade IV glioma (GBM) than WHO grade II and grade III glioma (Figure 1A and 1B). TCGA-GBM database show that mRNA level of LHPP was significantly lower in GBM, compared with normal brain tissue (Figure 1C). Furthermore, Kaplan-Meier survival analysis of TCGA_GBMLGG and CGGA_mRNAseq database manifested that glioma patients with abnormal low mRNA level of LHPP exhibited a conspicuous poor overall survival than those patients with high mRNA level of LHPP (Figure 1D and 1E).
Figure 1.
Expression profile of LHPP in glioma and the clinical relevance. A. mRNA level of LHPP in different WHO grade based on TCGA_GBMLGG database. B. RNA level of LHPP in different WHO grade based on CGGA_mRNAseq database. C. mRNA level of LHPP between GBM and normal brain tissue (NBT) based on TCGA_GBM database. D. Kaplan-Meier curves of survival rate of glioma patients with high LHPP mRNA level and low LHPP mRNA level based on TCGA_GBMLGG database. E. Kaplan-Meier curves of survival rate of glioma patients with high LHPP mRNA level and low LHPP mRNA level based on CGGA_mRNAseq database. F. Representative immunohistochemistry staining of LHPP on glioma tissues microarrays (×400). G. Composition bar chart of LHPP immunohistochemistry staining score in different WHO grade. H. Kaplan-Meier curves of survival rate of glioma patients with high LHPP immunohistochemistry staining score and low LHPP immunohistochemistry staining score. Score 0 and 1 were grouped and considered as low expression, score 2 and 3 were grouped and considered as high expression. I. Western blot detection of LHPP in 10 paired glioblastoma tissue (T) and adjacent brain tissue (N). J. Western blot detection of LHPP in GBM cell lines and HA. The data were expressed as the mean ± SEM (***P<0.0001).
Next, we conducted immunohistochemistry (IHC) staining with a glioma tissue microarray slide, which containing 94 cases of glioma samples. We identified 20 cases classified score 0, 39 cases classified score 1, 29 cases classified score 2, and 6 cases classified score 3. The cases of score 0 and score 1 were defined as IHC low score, and cases of score 2 and score 3 were defined as IHC high score. The IHC staining scoring standard and schematic diagrams were shown (Figure 1F). We analyzed the IHC staining score and proportion of samples at different WHO grade, and we found that there was a tendency of low LHPP staining score in grade IV glioma (GBM), comparing with grade II and grade III gliomas (Figure 1G). In addition, Kaplan-Meier analysis showed significant correlation of LHPP expression with patient survival, the median survival of high LHPP IHC score was significantly higher than that of low LHPP IHC score (Figure 1H).
We also assessed LHPP protein expression difference between 10 glioblastoma tumor tissue (T) and corresponding adjacent brain tissue (N) by western blot. The result showed that the protein level of LHPP in six out of ten glioblastoma tissues were remarkably decreasing comparing with the corresponding adjacent brain tissues (Figure 1I). Furthermore, protein expression level of LHPP in 6 frequently-used glioblastoma cell lines (U87MG, U118MG, U251MG, T98G, A172, and LN229) and human astrocytes were also detected by western blot. The result showed that LHPP protein expression level was distinctly less in U87MG, U251, T98G, A172 than human astrocytes. While, LHPP protein expression level is approximate between U118MG and human astrocytes, and obviously higher in LN229 than human astrocytes (Figure 1J).
In sum, there was a tendency of decreasing expression level of LHPP in glioblastoma, and this tendency is correlated with a poor prognosis in glioma.
LHPP inhibits the growth of glioblastoma in vitro and in vivo
To explore the effect of LHPP on glioblastoma, we selected U87MG, which show significantly lower protein expression level of LHPP than human astrocytes, and U118MG, which show approximate protein expression level of LHPP with human astrocytes, to conduct the following experiments. Protein expression level of LHPP in negative cells, cells transfected with empty vector plasmids, and cells transfected with LHPP plasmids were detected by western blot, the result indicated a satisfactory overexpression efficiency in U87MG and U118MG cells (Figure S2A and S2B).
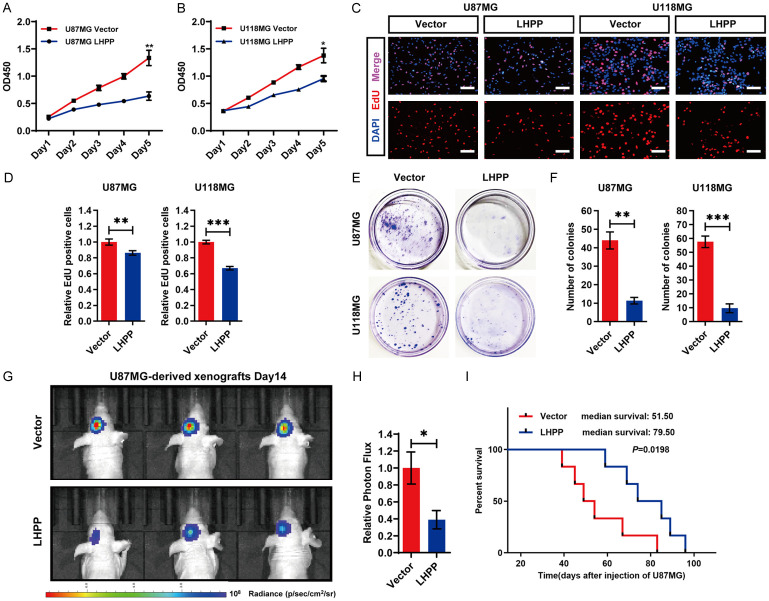
Firstly, we evaluated the effect of LHPP on U87MG and U118MG cells’ growth in vitro. CCK-8 assay was performed and the cell growth curve was drawn, the result indicated that LHPP-overexpression in U87MG and U118MG significantly inhibited the increase of cell number in vitro (Figure 2A and 2B). Then, EdU was used to label the proliferating cells (Figure 2C), the quantitative analysis revealed that LHPP-overexpression significantly decreased the ratio of EdU-positive cells in U87MG and U118MG cells (Figure 2D). In addition, cell clone formation assay was also performed, colonies were stained by crystal violet (Figure 2E). The statistical analysis showed that LHPP-overexpression obviously reduced the number of clones in U87MG and U118MG (Figure 2F). These results suggested that LHPP-overexpression inhibits the growth of glioblastoma in vitro.
Figure 2.
Overexpression of LHPP inhibits glioblastoma progression in vitro and in vivo. A and B. Cell growth curve of U87MG and U118MG cells after transfected with LHPP plasmids or empty vector, detected by the CCK-8 assay (n=4). C. EdU-labeled proliferating cells in U87MG and U118MG cells after transfected with LHPP plasmids or empty vector (×400). D. Statistical result of the EdU-Positive cells ratio in U87MG and U118MG cells after transfected with LHPP plasmids or empty vector (n=12, 3 transfection batches, each batch with 4 replicates). E. Cell clone formation of U87MG and U118MG cells after transfected with LHPP plasmids or empty vector, colonies were stained by crystal violet. F. Statistical result of the number of colonies in U87MG and U118MG cells after transfected with LHPP plasmids or empty vector (n=3). G. Representative in vivo IVIS Luc fluorescence image of xenografts mice at 14th day. H. Statistical result of photon flux in xenografts mice at 14th day (n=9). I. Kaplan-Meier curves of survival rate of xenografts mice (n=6). The data were expressed as the mean ± SEM (*P<0.05, **P<0.001, ***P<0.0001).
Then, in vivo study was performed by Intracranial glioma xenograft model. U87MG-Luc LHPP-overexpression, and U87MG-Luc Vector cells were transplanted into the skull of nude mice and allowed to develop for 14 days. Representative in vivo IVIS Luc fluorescence images at 14th day and the quantitative analysis of xenografts in mice suggested that the intracerebral tumor average size was significant smaller in nude mice which were transplanted with U87MG-Luc LHPP-overexpression cells (Figure 2G and 2H). The Kaplan-Meier analysis showed that the median survival time were significant longer in the nude mice which were transplanted with U87MG-Luc LHPP-overexpression cells (Figure 2I). These results suggested that LHPP-overexpression inhibits the growth of glioblastoma in vivo.
In sum, LHPP significantly inhibits the growth of glioblastoma in vitro and in vivo.
LHPP impedes the energy metabolism by restraining the glycolysis and respiration in glioblastoma
Metabolic reprogramming is a prominent feature of cancer cells, which mainly represented as enhanced aerobic glycolysis cooperating with tricarboxylic acid cycle, for providing efficient energy for the rapid growth of cancer cells in different tumor microenvironments [22,23].
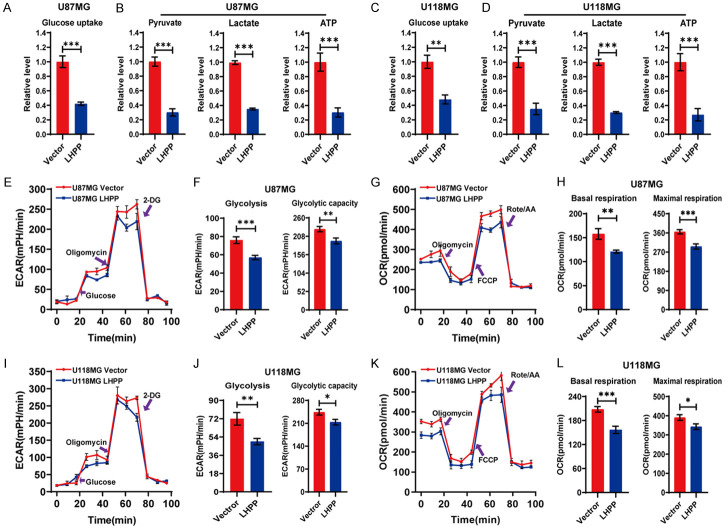
Under normoxic conditions, cancer cells would increase their glucose uptake, and preferentially utilize glucose through aerobic glycolysis, which allowing cancer cells to produce the energy more quickly and efficiently, and it is critical for cancer cells self-progression [24]. In this study, we analyzed whether LHPP affects the aerobic glycolysis. The level of glucose uptake was tested, and we found that the level of glucose uptake was significantly decreased after LHPP-overexpression in U87MG and U118MG cells (Figure 3A and 3C). Then, we tested the level of related metabolites in glycolysis and the production of ATP. The results revealed that the level of pyruvate, lactate, and the production of ATP were all significantly decreased in LHPP-overexpression U87MG and U118MG cells (Figure 3B and 3D). After that, we performed an extracellular acidification rate (ECAR) assay, which reflect the cancer cell’s state of overall glycolytic flux in vitro. The results suggested that LHPP-overexpression conspicuously reduced the extracellular acidification rate during the stage of glycolysis after glucose feeding and oligomycin feeding in U87MG and U118MG cells (Figure 3E and 3I). The quantitative analysis revealed that the level of glycolysis and glycolytic capacity were both significantly decreased in LHPP-overexpression U87MG and U118MG cells (Figure 3F and 3J). These results indicated that LHPP could impedes that aerobic glycolysis in glioblastoma cells.
Figure 3.
LHPP impedes energy metabolism in glioblastoma cells. A. Glucose uptake level was detected, LHPP significantly reduced the Glucose uptake in U87MG cells (n=4). B. Pyruvate, lactate, and ATP level in U87MG cells were measured, LHPP significantly reduced these metabolites in U87MG cells (n=4). C. Glucose uptake level was detected, LHPP significantly reduced the Glucose uptake in U118MG cells (n=4). D. Pyruvate, lactate, and ATP level in U118MG cells were measured, LHPP significantly reduced these metabolites in U87MG cells (n=4). E. Extracellular acidification rate (ECAR) assay by Seahorse XF96 Extracellular Flux Analyzer in U87MG cells. F. Statistical analysis of Glycolysis and Glycolytic capacity in U87MG cells after transfection with LHPP plasmids or empty vector (n=4). G. Oxygen consumption rate (OCR) assay by Seahorse XF96 Extracellular Flux Analyzer in U87MG cells. H. Statistical analysis of Basal respiration and Maximal respiration in U87MG cells after transfection with LHPP plasmids or empty vector (n=4). I. Extracellular acidification rate (ECAR) assay by Seahorse XF96 Extracellular Flux Analyzer in U118MG cells. J. Statistical analysis of Glycolysis and Glycolytic capacity in U118MG cells after transfection with LHPP plasmids or empty vector (n=4). K. Oxygen consumption rate (OCR) assay by Seahorse XF96 Extracellular Flux Analyzer in U118MG cells. L. Statistical analysis of Basal respiration and Maximal respiration in U118MG cells after transfection with LHPP plasmids or empty vector (n=4). The data were expressed as the mean ± SEM (*P<0.05, **P<0.001, ***P<0.0001).
Although aerobic glycolysis is enhanced in cancer cells, emerging studies suggested that cancer cells still dependent on TCA cycle for energy production under uncontrolled expression of oncogenes and tumor suppressor genes [25,26]. To explore whether LHPP interferes the TCA cycle, the oxygen consumption rate (OCR) assay was performed to explore effect of LHPP on the mitochondrial oxidative respiration. The results showed that LHPP-overexpression significantly decreased the oxygen consumption rate during mitochondrial oxidative respiration stage in U87MG and U118MG cells (Figure 3G and 3K). The quantitative analysis revealed that the level of basal respiration and maximal respiration were both dramatically decreased in LHPP-overexpression U87MG and U118MG cells (Figure 3H and 3L). These results indicated that LHPP-overexpression could impedes the respiration in glioblastoma cells.
In sum, LHPP impedes the energy metabolism by restraining the glycolysis and respiration in glioblastoma cells.
LHPP down-regulates the protein level and enzymatic activity of pyruvate kinase isoform M2 (PKM2) in glioblastoma
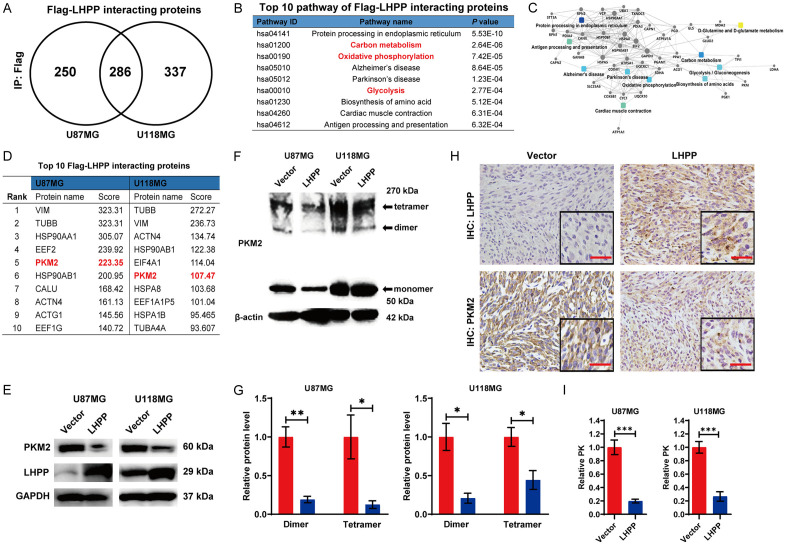
To investigate the underlying mechanism how LHPP restrained the energy metabolism, we performed mass-spectrometric assay to study the LHPP interacting proteins in glioblastoma cells. Anti-Flag antibody was used to precipitate Flag-LHPP. We detected 536 and 623 Flag-LHPP interacting proteins in LHPP-overexpression U87MG and U118MG cells respectively, and 286 proteins were coincident Flag-LHPP interacting proteins (Figure 4A). KEGG pathway analysis of these coincident proteins showed that 3/10 among the top 10 pathways were related with energy metabolic process, including Carbon metabolism, Oxidative phosphorylation, and Glycolysis (Figure 4B). Then protein-protein interaction network assay was performed, we found the proteins which involving in both glycolysis and carbon metabolism were PGAM1, PGK1, LDH, GAPDH, TPI1, and PKM2, respectively (Figure 4C). Among these six proteins, the PKM2 is the only Flag-LHPP interacting protein which ranked top 10 either in U87MG, or U118MG cells (Figure 4D). Then, we analyzed the expression level of PKM2 by western blot, and we found that the protein level of PKM2 was significant down-regulated in LHPP-overexpression U87MG and U118MG cells (Figure 4E). These results suggested that the PKM2 may be the potential target for LHPP-overexpression inhibiting energy metabolism glioblastoma cells.
Figure 4.
LHPP down-regulated the protein level and enzymatic activity of PKM2. A. Venn diagram of Flag-LHPP interacting proteins in LHPP-overexpression U87MG and U118MG cells, and the coincident proteins anti-Flag antibody was used to pull down the Flag-LHPP complex. B. Top 10 KEGG pathways of coincident Flag-LHPP interacting proteins in LHPP-overexpression U87MG and U118MG cells. C. Protein-protein interaction network of coincident Flag-LHPP interacting proteins in LHPP-overexpression U87MG and U118MG cells. D. Top 10 Flag-LHPP interacting proteins in U87MG and U118MG cells. E. Western blot assay for protein expression level of PKM2 in U87MG and U118MG cells after LHPP plasmids or empty vector were transfected. F. Western blot detect the protein expression level of monomeric, dimeric, and tetrameric PKM2 in U87MG and U118MG cells after LHPP plasmids or empty vector were transfected. G. Statistical analysis of relative protein expression level of dimeric and tetrameric PKM2 in U87MG and U118MG cells after LHPP plasmids or empty vector were transfected (n=3). H. Immunohistochemical staining of LHPP and PKM2 in paraffin sections of the brain of the tumor-burdened mice (×400). I. Pyruvate kinase activity (PK) was detected in U87MG and U118MG cells after LHPP plasmids or empty vector were transfected, and the statistical analysis of relative PK (n=4). The data were expressed as the mean ± SEM (*P<0.05, **P<0.001, ***P<0.0001).
PKM2 plays a role of converting phosphoenolpyruvate (PEP) to pyruvate at the final step of glycolysis [27]. There are three dynamic populations of PKM2 existing in mammalian cell, including monomer, dimer and tetramer, but only dimeric and tetrameric PKM2 exert pyruvate kinase activity [28]. The monomeric PKM2 is an inactive state. The dimeric PKM2 prompts the glucose-derived carbon to the direction of glycolysis. While, the tetrameric PKM2 prompts glucose-derived carbon to the direction of respiratory chain [29]. The previous results revealed that the glycolysis and respiratory were both inhibited after LHPP overexpression, which also suggested that PKM2 may be the crucial target. Subsequently, we analyzed the effect of LHPP on the dynamic populations of PKM2 in glioblastoma cells. Intracellular dynamic populations of PKM2 were preserved via formaldehyde cross-linking treatment. Then, we detected the protein level of different PKM2 dynamic populations by western blot (Figure 4F). Quantitative analysis revealed that dimeric and tetrameric PKM2 were both significant down-regulated in LHPP-overexpression U87MG and U118MG cells (Figure 4G). In addition, IHC staining of PKM2 in paraffin sections of intracranial glioma xenograft model was performed, and the result also suggested that LHPP-overexpression down-regulated the protein level of PKM2 (Figure 4H). Finally, we measured the pyruvate kinase activity (PK), the result showed that the PK value was evidently decreasing in LHPP-overexpression U87MG and U118MG cells (Figure 4I).
In sum, LHPP down-regulates the protein level and enzymatic activity of PKM2 in glioblastoma cells.
LHPP does not regulate the transcription level of PKM2, nor dose obviously reprogram energy metabolic genome transcription
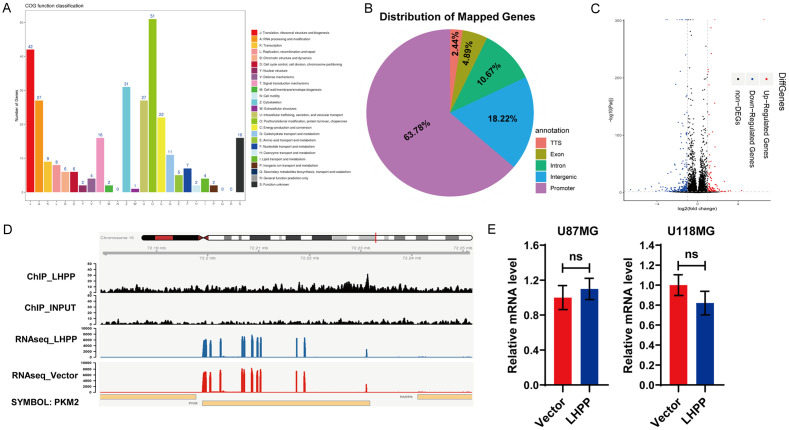
The previous studies have demonstrated that LHPP down-regulated the protein level of PKM2. Since COG function classification result of mass-spectrometric analysis showed that LHPP interacting proteins were involved in Transcription, Chromatin structure and dynamics (Figure 5A). This result prompted us to investigate whether the reduced protein level of PKM2 was depend on declining gene transcription level, and whether LHPP directly reprogram the energy metabolic genome transcription.
Figure 5.
LHPP binding regions on chromosome are independent of PKM2 transcription and metabolic genome reprogramming. A. COG function classification of coincident Flag-LHPP interacting proteins in Figure 4A. B. Pie chart display the genomic distribution of Flag-LHPP binding sites compared to background. C. Volcano plot of differentially expressed genes (DEGs) based on the result of RNAseq. D. Alignment data of LHPP ChIPseq and RNAseq of U87MG OE LHPP cells near the LHPP and PKM2 locus. E. mRNA level of PKM2 was detected by qRT-PCR in U87MG and U118MG cells after LHPP plasmids or empty vector were transfected, the statistical analysis show there were no significant difference (n=4). The data were expressed as the mean ± SEM (ns P>0.05).
To clarify above questions, we performed a ChIPseq and RNAseq integrated analysis. Anti-Flag antibody was used to precipitate Flag-LHPP in LHPP-overexpression U87MG cells, and we identified 455 regions of which bound by Flag-LHPP, that corresponded to mostly promoter regions (63.78%), follow by intergenic regions (18.22%), intron regions (10.67%), exon regions (4.89%), and TTS regions (2.44%) (Figure 5B). However, enrichment results of Flag-LHPP bound regions indicated that LHPP does not directly participate in energy metabolic genome reprogramming, and there were not LHPP-dependent co-factors which regulate PKM2 activation. As to RNAseq, there were 534 differentially expressed genes (DEGs), include 124 up-regulated genes and 410 down-regulated genes in LHPP-overexpression U87MG cells, comparing to control cells (Figure 5C). Visualization result of ChIPseq and RNAseq reads at the located locus of LHPP and PKM2 showed that LHPP do not obviously located the locus of PKM2 (Figure 5D). In addition, qRT-PCR identified that LHPP-overexpression indeed did not obviously regulate LHPP mRNA level in U87MG and U118MG cells (Figure 5E).
In sum, LHPP does not regulate the transcription level of PKM2, nor does obviously reprogram the energy metabolic genome transcription. The reduced protein level of PKM2 caused by LHPP-overexpression in U87MG and U118MG cells is independent of transcription level.
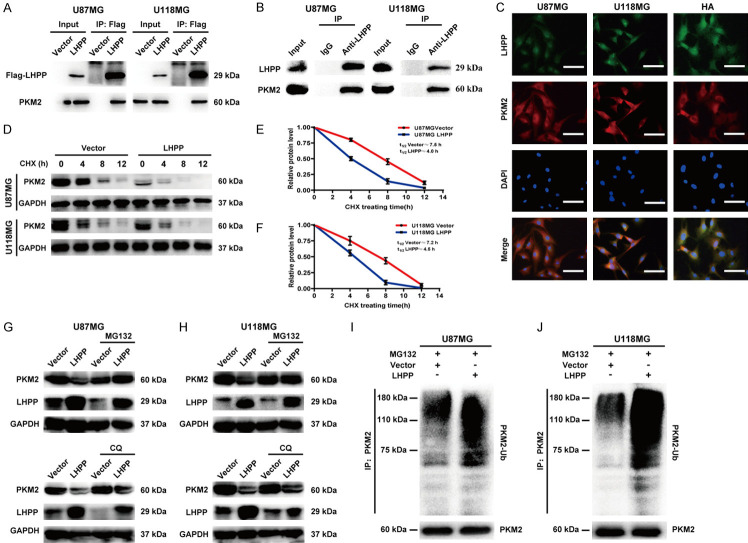
LHPP interacts with PKM2 and induces ubiquitin-mediated degradation of PKM2
Due to LHPP does not regulate the transcription level of PKM2, we speculated that the reduced protein level of PKM2 caused by LHPP-overexpression may be related with post-transcriptional modification. The mass-spectrometric analysis indicated that LHPP could interacts with PKM2 (Figure 4C). We performed Co-IP assay to verify the interaction between LHPP and PKM2, exogenous LHPP was precipitated with anti-Flag antibody in LHPP-overexpression U87MG and U118MG cells, and we detected PKM2 (Figure 6A); Meanwhile, endogenous LHPP was precipitated with anti-LHPP antibody in U87MG and U118MG cells, we also detected PKM2 (Figure 6B). In addition, we performed immunofluorescence assay for analyzing the interaction between LHPP and PKM2 based on protein spatial position. We found that LHPP is low expression and PKM2 is relatively high expression in cytoplasm of U87MG and U118MG cells. However, LHPP is high expression and PKM2 is relatively low expression in cytoplasm of HA cells. LHPP and PKM2 had colocalization in cytoplasm, especially in cytoplasm of HA cells (Figure 6C). Furthermore, immunofluorescence assay for Flag-LHPP and PKM2 was also performed by using LHPP-overexpression U87MG and U118MG cells, and we found that Flag-LHPP and PKM2 also had obvious colocalization in cytoplasm (Figure S3). Thus, these results confirmed that LHPP can interact with PKM2.
Figure 6.
LHPP interacts with PKM2 and induces ubiquitin-mediated degradation of PKM2. A. Co-immunoprecipitation of exogenous LHPP and PKM2 in LHPP plasmids and vector transfected U87MG and U118MG cells, exogenous LHPP was pulled down by using anti-Flag antibody, and PKM2 was detected. B. Co-immunoprecipitation of endogenous LHPP and PKM2 in U87MG and U118MG cells, endogenous LHPP was pulled down by using anti-LHPP antibody, and PKM2 was detected. C. Immunofluorescence colocalization assay of LHPP and PKM2 in U87MG, U118MG and HA cells. Green fluorescence stands for LHPP, red fluorescence stands for PKM2, and blue fluorescence stands for nucleus (×400). D. Time course analysis of PKM2 degradation in U87MG or U118MG cells after LHPP plasmids or empty vector transfected and CHX (100 μg/mL) treated in was detected by western blot. E and F. Protein level curve for PKM2 based on time course analysis in U87MG and U118MG cells (n=3). G and H. Protein expression level of PKM2 in U87MG or U118MG cells after LHPP plasmids or empty vector transfected and incubated with MG132 (5 μM) or CQ (20 μM) for 6 hours was detected by western blot. I and J. U87MG or U118MG cells transfected with LHPP plasmids or empty vector were treated with MG132 (10 μM) for 6 hours. Ubiquitination level of PKM2 were detected by immunoprecipitation assay.
To explored whether LHPP interacts with PKM2 interfered protein stability of PKM2, we used a de novo protein synthesis inhibitor, CHX, to treat the cells, and the protein level of PKM2 was detected by western blot (Figure 6D). Statistical analysis indicated that the protein half-life of PKM2 in cells of which transfected with LHPP and CHX (4.0 hours in U87MG, and 4.5 hours in U118MG) was significant shorted than that in the cells of which treated with empty vector and CHX (7.5 hours in U87MG, and 7.2 hours in U118MG) (Figure 6E and 6F). This result suggested that LHPP interfered the protein stability of PKM2 and promoted degradation of PKM2 in U87MG and U118MG cells.
Previous studies demonstrated that PKM2 could be degraded via ubiquitin-mediated proteolytic pathway [30] or autophagy-lysosome proteolytic pathway [31]. In this study, we used MG132 (a proteasome blocker), and CQ (a lysosome inhibitor) to block the ubiquitin-mediated proteolytic pathway and autophagy-lysosome proteolytic pathway, respectively. We found that MG132, but not CQ, reversed the decreasing protein level of PKM2 in LHPP-overexpression U87MG and U118MG cells (Figure 6G and 6H). After that, in vitro ubiquitination activity assay was performed to explore role of ubiquitin in LHPP-overexpression mediated protein degradation of PKM2 in U87MG and U118MG cells. Cells were preincubated with MG132, and transfected with empty vector or LHPP plasmids. Then, the cell lysates were incubated with anti-PKM2 antibody, and immunoprecipitated proteins were assessed by using an anti-ubiquitin antibody. The results revealed that the ubiquitination of PKM2 was augmented in LHPP-overexpression U87MG and U118MG cells (Figure 6I and 6J). Taken together, these results suggested that the protein degradation of PKM2 caused by LHPP-overexpression was through ubiquitin-mediated proteolytic pathway.
In sum, LHPP interacts with PKM2 and induces ubiquitin-mediated degradation of PKM2.
Restoration of PKM2 reversed the inhibition of energy metabolism and glioblastoma growth induced by LHPP
We performed experiments in vitro and in vivo to explore whether restoring the PKM2 protein expression could reverse the inhibited cancer growth and restrained energy metabolism in LHPP-overexpression U87MG and U118MG cells. Glucose uptake assay showed that the level of glucose uptake was return to normal even slightly higher after restoration of PKM2 in LHPP-overexpression U87MG and U118MG cells (Figure 7A). The ECAR and OCR assay were performed, the curve showed that the restoration of PKM2 could resist LHPP-overexpression caused decreasing ECAR and OCR level in U87MG and U118MG cells (Figure 7B and 7C). The production of pyruvate, lactate, and ATP were all recovered after restoration of PKM2 expression in LHPP-overexpression U87MG and U118MG cells (Figure S4A and S4B). In addition, statistical analysis of ECAR and OCR both revealed that restoration of PKM2 regained the normal, even slightly increasing level of aerobic glycolysis and respiration in LHPP-overexpression U87MG and U118MG cells (Figure S4C-F). These results indicated that restoration of PKM2 expression could reverse the LHPP-overexpression caused inhibition of glycolysis and respiration in glioblastoma cells. As to cancer growth, cell clone formation assay was performed (Figure 7D), the quantitative analysis showed that after restoration of PKM2 expression, the clone number in LHPP-overexpression U87MG and U118MG cells were not significantly different from those cells transfected with empty vector (Figure 7E). In vivo, intracranial glioma xenograft model was performed, the representative images in vivo IVIS Luc fluorescence image at 14th day of mice in vector group, LHPP group, and LHPP plus PKM2 group were shown (Figure 7F). The quantitative analysis of photon flux in mice suggested the cancer cell growth in vivo was recovered after restoration of PKM2 expression in LHPP-overexpression U87MG-Luc cells (Figure 7G).
Figure 7.
Restoration of PKM2 reversed restrained energy metabolism and glioblastoma growth inhibition caused by LHPP-overexpression. A. Glucose uptake was detected in glioblastoma cells transfected with empty vector, LHPP plasmids, and LHPP plus PKM2 plasmids, and the statistical analysis showed that restoration of PKM2 reversed the decreasing level of glucose uptake caused by LHPP in U87MG or U118MG cells (n=4). B. Extracellular acidification rate (ECAR) assay by Seahorse XF96 Extracellular Flux Analyzer in U87MG and U118MG cells after transfected with empty vector, LHPP plasmids, and LHPP plus PKM2 plasmids. C. Oxygen consumption rate (OCR) assay by Seahorse XF96 Extracellular Flux Analyzer in U87MG and U118MG cells after transfected with empty vector, LHPP plasmids, and LHPP plus PKM2 plasmids. D. Cell clone formation assay in U87MG and U118MG cells after transfected with empty vector, LHPP plasmids, and LHPP plus PKM2 plasmids. E. Statistical result of the number of colonies in U87MG and U118MG cells after transfected with empty vector, LHPP plasmids, and LHPP plus PKM2 plasmids (n=4). F. Representative in vivo IVIS Luc fluorescence image of xenografts mice at 14th day. G. Statistical result of photon flux in xenografts mice at 14th day (n=8). The data were expressed as the mean ± SEM (*P<0.05, **P<0.001, ***P<0.0001, ns P>0.05).
In sum, restoration of PKM2 could effectively reverse the inhibition of energy metabolism and cancer cell growth caused by LHPP-overexpression in glioblastoma.
Discussion
In this study, we focused on LHPP, which had been proved to be a novel tumor suppressor in hepatocellular carcinoma by Hindupur SK [15], and subsequent researches indicated that LHPP maybe a tumor suppressor in multisystem tumors [16-20]. While, the role of LHPP in glioma is still unclear. Base on the TCGA and CGGA databases, and preliminary experimental study, we demonstrated that that LHPP is trend to high expression in normal brain tissue, but decreasing or absent in glioblastoma, and the expression level of LHPP was correlated with the median survival of glioma patients. The laboratory studies identified that LHPP-overexpression significantly inhibited the growth of glioblastoma cells in vitro and in vivo. Based on these results, we speculated that LHPP maybe a tumor suppressor in glioblastoma as well.
Cancer cells demand a high-efficiency energy supplying for maintaining rapidly proliferation and tumor growth. Energy metabolism reprogramming is a hallmark of cancer, mainly for enhanced aerobic glycolysis, in coordination with respiration under different oxygen conditions, and both are important for rapid cancer cell growth [23,32]. Our study revealed that glycolysis was suppressed in LHPP-overexpression U87MG and U118MG cells, which is manifested that the extracellular acidification rate was slowing, and the production of pyruvate, lactate, and ATP were decreasing, as well as glucose uptake level was restrained significantly. Meanwhile, the respiration level was also depressed in LHPP-overexpression U87MG and U118MG cells, which is manifested that the basal respiration and maximal respiration were reduced significantly. These results indicated that LHPP impedes energy metabolism in glioblastoma.
For the underlying mechanism of LHPP impedes glycolysis and respiration in glioblastoma, the mass-spectrometric analysis revealed that top 10 enriched pathways of LHPP interacting proteins were involved in energy metabolic process, including carbon metabolism, oxidative phosphorylation, and glycolysis. This result suggested that LHPP impedes energy metabolism in glioblastoma may via regulating these interacting proteins. Western blot assay identified that PKM2, one of LHPP interacting proteins which is the top ranked among the coincident LHPP interacting proteins in U87MG and U118MG cells, was significantly decreased after LHPP-overexpression in U87MG and U118MG cells. PKM2 is a rate-limiting enzyme which converts the phosphoenolpyruvate (PEP) to pyruvate in glycolysis, and it is a key checkpoint for energy metabolism [27,28]. Interestingly, the function of PKM2 on glycolysis and respiration is complicated. Y Shang reported that down-regulated PKM2 caused by CHIP/Stub1 lead to decreased glycolysis, but increased respiration [30]. While, Tong Li reported that knockdown of PKM2 in the lung cancer cells would both inhibit glycolysis and respiration [33]. PKM2 has three dynamic populations in mammalian cells, respectively are monomer, dimer, and tetramer, and they exhibit different functions [34]. Monomeric PKM2 is an inactive state. Dimeric PKM2 has low catalytic activity, which prefer to prompt glucose-derived carbon to the direction of glycolysis. While, tetrameric PKM2 has high catalytic activity, which prefer to prompt glucose-derived carbon to the direction of respiratory chain [29,35]. The variation difference between dimeric and tetrameric PKM2 may help to explain the above-mentioned discrepancy. Hence, protein level of dimeric and tetrameric PKM2 was detected using formaldehyde Cross-linking protein samples, and we found that dimeric and tetrameric PKM2 were both down-regulated in LHPP-overexpression U87MG and U118MG cells. These results just explained why LHPP-overexpression induced codirectional inhibition of glycolysis and respiration.
The underlying mechanism by which LHPP regulates PKM2 is unclear. Previous studies have demonstrated that the network of PI3K-AKT-mTOR, hypoxia-inducible factor 1 (HIF-1), c-Myc are momentous factors in regulating the transcription of PKM2 [36-38]. In our study, the Co-IP assay and immunofluorescence assay confirmed that LHPP could interact with PKM2, and the COG function classification indicated that LHPP interacting proteins were involved in Transcription, Chromatin structure and dynamics. These results led us to wonder whether LHPP could cause changes in PKM2 through direct gene transcription regulation or direct action. We explored the function of LHPP on metabolic genome programming and the transcription of PKM2 by ChIPseq and RNAseq integrated analysis. However, LHPP do not significantly bound to the elements which could regulate the transcription of PKM2, and LHPP do not obviously change the metabolic genome transcription of which regulate glycolysis and respiration.
Since the reduced protein level of PKM2 is independent on gene transcription, we guessed that post-transcriptional modification might be responsible for the decreased protein level of PKM2. Previous studies demonstrated that PKM2 could be degraded via ubiquitin-mediated proteolytic pathway or autophagy-lysosome proteolytic pathway [30]. We treated the cells with CHX, a protein synthesis inhibitor, the result showed that the degradation half-life of PKM2 in LHPP-overexpressed U87MG and U118MG cells were both shortened significantly, which means the degradation rate of PKM2 was accelerated by LHPP-overexpression. Then, MG132, a proteasome inhibitor, and CQ, a lysosome inhibitor, were used to block the ubiquitin-mediated proteolytic pathway and autophagy-lysosome proteolytic pathway, respectively. We found that only MG132 could apparently rescued the protein level of PKM2 in LHPP-overexpression U87MG and U118MG cells. Furthermore, ubiquitination activity assay revealed that LHPP-overexpression augmented ubiquitination of PKM2. Taken together, LHPP induced ubiquitin-mediated degradation of PKM2. We performed rescue assay for PKM2 in vitro and in vivo. The results indicated that restoration of PKM2 expression could effectively reverse the restrained energy metabolic process, include glycolysis and respiration. Meanwhile, the glioblastoma growth in vitro and in vivo were all regained after restoration of PKM2 expression in LHPP-overexpression glioblastoma cells.
In summary, we uncovered that LHPP induces ubiquitin-mediated degradation of PKM2, and then impedes the energy metabolic process, including glycolysis and respiration, which finally leads to significant growth inhibition of glioblastoma in vitro and in vivo. This is the first work to uncover the function and metabolic correlation mechanism of LHPP in glioblastoma, and lays a foundation for the future diagnosis and treatment of glioblastoma.
Acknowledgements
This work was supported by a grant from the National Natural Science Foundation of China (Grant No. 81573774), and the Military Medical Science Research Project (16CXZ001). The study was approved by the ethics committee of the Southern Medical University and The Seventh Medical Center of General Hospital of PLA. The collection and using of human glioblastoma tissues in this study was in accord with the ethical standards established by ethics committee of The Seventh Medical Center of General Hospital of PLA. All animal experiments were approved by the Institutional Animal Care and Use Committee of Southern Medical University and The Seventh Medical Center of General Hospital of PLA.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Ostrom QT, Gittleman H, de Blank PM, Finlay JL, Gurney JG, McKean-Cowdin R, Stearns DS, Wolff JE, Liu M, Wolinsky Y, Kruchko C, Barnholtz-Sloan JS. American brain tumor association adolescent and young adult primary brain and central nervous system tumors diagnosed in the United States in 2008-2012. Neuro Oncol. 2016;18(Suppl 1):i1–i50. doi: 10.1093/neuonc/nov297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359:492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- 3.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, Curschmann J, Janzer RC, Ludwin SK, Gorlia T, Allgeier A, Lacombe D, Cairncross JG, Eisenhauer E, Mirimanoff RO European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups; National Cancer Institute of Canada Clinical Trials Group. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 4.Chen J, Li Y, Yu TS, McKay RM, Burns DK, Kernie SG, Parada LF. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488:522–526. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, Brennan C, Chin L, DePinho RA, Cavenee WK. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 6.Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC, Ludwin SK, Allgeier A, Fisher B, Belanger K, Hau P, Brandes AA, Gijtenbeek J, Marosi C, Vecht CJ, Mokhtari K, Wesseling P, Villa S, Eisenhauer E, Gorlia T, Weller M, Lacombe D, Cairncross JG, Mirimanoff RO European Organisation for Research and Treatment of Cancer Brain Tumour and Radiation Oncology Groups; National Cancer Institute of Canada Clinical Trials Group. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10:459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 7.Carlsson SK, Brothers SP, Wahlestedt C. Emerging treatment strategies for glioblastoma multiforme. EMBO Mol Med. 2014;6:1359–1370. doi: 10.15252/emmm.201302627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.von Neubeck C, Seidlitz A, Kitzler HH, Beuthien-Baumann B, Krause M. Glioblastoma multiforme: emerging treatments and stratification markers beyond new drugs. Br J Radiol. 2015;88:20150354. doi: 10.1259/bjr.20150354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reifenberger G, Wirsching HG, Knobbe-Thomsen CB, Weller M. Advances in the molecular genetics of gliomas - implications for classification and therapy. Nat Rev Clin Oncol. 2017;14:434–452. doi: 10.1038/nrclinonc.2016.204. [DOI] [PubMed] [Google Scholar]
- 10.Seal US, Binkley F. An inorganic pyrophosphatase of swine brain. J Biol Chem. 1957;228:193–199. [PubMed] [Google Scholar]
- 11.Ek P, Pettersson G, Ek B, Gong F, Li JP, Zetterqvist O. Identification and characterization of a mammalian 14-kDa phosphohistidine phosphatase. Eur J Biochem. 2002;269:5016–5023. doi: 10.1046/j.1432-1033.2002.03206.x. [DOI] [PubMed] [Google Scholar]
- 12.Cui L, Gong X, Tang Y, Kong L, Chang M, Geng H, Xu K, Wang F. Relationship between the LHPP gene polymorphism and resting-state brain activity in major depressive disorder. Neural Plast. 2016;2016:9162590. doi: 10.1155/2016/9162590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cui L, Wang F, Yin Z, Chang M, Song Y, Wei Y, Lv J, Zhang Y, Tang Y, Gong X, Xu K. Effects of the LHPP gene polymorphism on the functional and structural changes of gray matter in major depressive disorder. Quant Imaging Med Surg. 2020;10:257–268. doi: 10.21037/qims.2019.12.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neff CD, Abkevich V, Packer JC, Chen Y, Potter J, Riley R, Davenport C, DeGrado Warren J, Jammulapati S, Bhathena A, Choi WS, Kroeger PE, Metzger RE, Gutin A, Skolnick MH, Shattuck D, Katz DA. Evidence for HTR1A and LHPP as interacting genetic risk factors in major depression. Mol Psychiatry. 2009;14:621–630. doi: 10.1038/mp.2008.8. [DOI] [PubMed] [Google Scholar]
- 15.Hindupur SK, Colombi M, Fuhs SR, Matter MS, Guri Y, Adam K, Cornu M, Piscuoglio S, Ng CKY, Betz C, Liko D, Quagliata L, Moes S, Jenoe P, Terracciano LM, Heim MH, Hunter T, Hall MN. The protein histidine phosphatase LHPP is a tumour suppressor. Nature. 2018;555:678–682. doi: 10.1038/nature26140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hou B, Li W, Li J, Ma J, Xia P, Liu Z, Zeng Q, Zhang X, Chang D. Tumor suppressor LHPP regulates the proliferation of colorectal cancer cells via the PI3K/AKT pathway. Oncol Rep. 2020;43:536–548. doi: 10.3892/or.2019.7442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Y, Zhang X, Zhou X, Zhang X. LHPP suppresses bladder cancer cell proliferation and growth via inactivating AKT/p65 signaling pathway. Biosci Rep. 2019;39:BSR20182270. doi: 10.1042/BSR20182270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun W, Qian K, Guo K, Chen L, Xiang J, Li D, Wu Y, Ji Q, Sun T, Wang Z. LHPP inhibits cell growth and migration and triggers autophagy in papillary thyroid cancer by regulating the AKT/AMPK/mTOR signaling pathway. Acta Biochim Biophys Sin (Shanghai) 2020;52:382–389. doi: 10.1093/abbs/gmaa015. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Q, Xiong M, Liu J, Wang S, Du T, Kang T, Liu Y, Cheng H, Huang M, Gou M. Targeted nanoparticle-mediated LHPP for melanoma treatment. Int J Nanomedicine. 2019;14:3455–3468. doi: 10.2147/IJN.S196374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng J, Dai X, Chen H, Fang C, Chen J, Sun L. Down-regulation of LHPP in cervical cancer influences cell proliferation, metastasis and apoptosis by modulating AKT. Biochem Biophys Res Commun. 2018;503:1108–1114. doi: 10.1016/j.bbrc.2018.06.127. [DOI] [PubMed] [Google Scholar]
- 21.Vasilescu J, Guo X, Kast J. Identification of protein-protein interactions using in vivo cross-linking and mass spectrometry. Proteomics. 2004;4:3845–3854. doi: 10.1002/pmic.200400856. [DOI] [PubMed] [Google Scholar]
- 22.Ganapathy-Kanniappan S, Geschwind JF. Tumor glycolysis as a target for cancer therapy: progress and prospects. Mol Cancer. 2013;12:152. doi: 10.1186/1476-4598-12-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sajnani K, Islam F, Smith RA, Gopalan V, Lam AK. Genetic alterations in Krebs cycle and its impact on cancer pathogenesis. Biochimie. 2017;135:164–172. doi: 10.1016/j.biochi.2017.02.008. [DOI] [PubMed] [Google Scholar]
- 24.Lunt SY, Vander Heiden MG. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu Rev Cell Dev Biol. 2011;27:441–464. doi: 10.1146/annurev-cellbio-092910-154237. [DOI] [PubMed] [Google Scholar]
- 25.Anderson NM, Mucka P, Kern JG, Feng H. The emerging role and targetability of the TCA cycle in cancer metabolism. Protein Cell. 2018;9:216–237. doi: 10.1007/s13238-017-0451-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kerr EM, Gaude E, Turrell FK, Frezza C, Martins CP. Mutant Kras copy number defines metabolic reprogramming and therapeutic susceptibilities. Nature. 2016;531:110–113. doi: 10.1038/nature16967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dayton TL, Jacks T, Vander Heiden MG. PKM2, cancer metabolism, and the road ahead. EMBO Rep. 2016;17:1721–1730. doi: 10.15252/embr.201643300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wong N, De Melo J, Tang D. PKM2, a central point of regulation in cancer metabolism. Int J Cell Biol. 2013;2013:242513. doi: 10.1155/2013/242513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gui DY, Lewis CA, Vander Heiden MG. Allosteric regulation of PKM2 allows cellular adaptation to different physiological states. Sci Signal. 2013;6:pe7. doi: 10.1126/scisignal.2003925. [DOI] [PubMed] [Google Scholar]
- 30.Shang Y, He J, Wang Y, Feng Q, Zhang Y, Guo J, Li J, Li S, Wang Y, Yan G, Ren F, Shi Y, Xu J, Zeps N, Zhai Y, He D, Chang Z. CHIP/Stub1 regulates the Warburg effect by promoting degradation of PKM2 in ovarian carcinoma. Oncogene. 2017;36:4191–4200. doi: 10.1038/onc.2017.31. [DOI] [PubMed] [Google Scholar]
- 31.Wang C, Xu J, Yuan D, Bai Y, Pan Y, Zhang J, Shao C. Exosomes carrying ALDOA and ALDH3A1 from irradiated lung cancer cells enhance migration and invasion of recipients by accelerating glycolysis. Mol Cell Biochem. 2020;469:77–87. doi: 10.1007/s11010-020-03729-3. [DOI] [PubMed] [Google Scholar]
- 32.Corbet C, Feron O. Cancer cell metabolism and mitochondria: nutrient plasticity for TCA cycle fueling. Biochim Biophys Acta Rev Cancer. 2017;1868:7–15. doi: 10.1016/j.bbcan.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 33.Li T, Han J, Jia L, Hu X, Chen L, Wang Y. PKM2 coordinates glycolysis with mitochondrial fusion and oxidative phosphorylation. Protein Cell. 2019;10:583–594. doi: 10.1007/s13238-019-0618-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mazurek S, Boschek CB, Hugo F, Eigenbrodt E. Pyruvate kinase type M2 and its role in tumor growth and spreading. Semin Cancer Biol. 2005;15:300–308. doi: 10.1016/j.semcancer.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 35.Mazurek S. Pyruvate kinase type M2: a key regulator of the metabolic budget system in tumor cells. Int J Biochem Cell Biol. 2011;43:969–980. doi: 10.1016/j.biocel.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 36.Iqbal MA, Bamezai RN. Resveratrol inhibits cancer cell metabolism by down regulating pyruvate kinase M2 via inhibition of mammalian target of rapamycin. PLoS One. 2012;7:e36764. doi: 10.1371/journal.pone.0036764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun Q, Chen X, Ma J, Peng H, Wang F, Zha X, Wang Y, Jing Y, Yang H, Chen R, Chang L, Zhang Y, Goto J, Onda H, Chen T, Wang MR, Lu Y, You H, Kwiatkowski D, Zhang H. Mammalian target of rapamycin up-regulation of pyruvate kinase isoenzyme type M2 is critical for aerobic glycolysis and tumor growth. Proc Natl Acad Sci U S A. 2011;108:4129–4134. doi: 10.1073/pnas.1014769108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wong N, Ojo D, Yan J, Tang D. PKM2 contributes to cancer metabolism. Cancer Lett. 2015;356:184–191. doi: 10.1016/j.canlet.2014.01.031. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.