Abstract
Colorectal cancer (CRC) is regarded as the third most common cancer worldwide. Although Regorafenib as a receptor tyrosine kinase inhibitor (RTKI) disrupts tumor growth and angiogenesis in metastatic CRC (mCRC) patients, drug resistance leads to poor prognosis and survival. Integrin-β1 overexpression has been proposed to be the major player in this regard. Herein, the Regorafenib-resistant human colon cancer cell line (SW-48) was induced, and the Integrin-β1 gene expression, as well as apoptosis, was assessed through the combination of small interfering RNA (siRNA) targeting Integrin-β1 and Regorafenib/Dimethyldioctadecylammonium bromide (DDAB)-methoxy poly (ethylene glycol) (mPEG)-poly-ε-caprolactone (PCL) hybrid nanoparticles (HNPs). In the current study, Regorafenib-resistant SW-48 cell line was generated in which the Regorafenib half-maximal inhibitory concentration (IC50) for non-resistant and resistant cells was 13.5±1.5 µM and 55.1±0.8 µM, respectively. The results of DLS also demonstrated that the size and the charge of the HNPs were equal to 66.56±0.5 nm and +29.5±1.2 mv, respectively. In addition, the Integrin-β1 gene expression was significantly higher in resistant cells than in non-resistant ones (P<0.05). The siRNA/HNP complexes in combination with Regorafenib/HNPs were accordingly identified as the most effective treatment to decrease the Integrin-β1 gene expression and to enhance the apoptosis rate in resistant cells (P<0.001). Overall, the study indicated that combination therapy using siRNA/HNP and Regorafenib/HNPs complex could down-regulate the Integrin-β1 gene expression and consequently trigger apoptosis, and this may potentially induce drug sensitivity.
Keywords: Colorectal cancer, regorafenib, integrin-β1, SiRNA, lipid-polymer hybrid nanoparticle, apoptosis
Introduction
Colorectal cancer (CRC) is regarded as the third most prevalent cancer after lung and breast cancers and has been recently reported as the second common one in some countries. It has also been the second most cancer-related mortality globally after lung cancer in 2018 [1]. Concerning the CRC development, both genetic and environmental factors are involved; however, the highest percentage of cases with this cancer is deeply associated with environmental factors. CRCs occur sporadically in the majority of cases, in which chromosomal instability (CIN), microsatellite instability (MSI), and epigenetic alterations constitute three major factors in the carcinogenic process. Furthermore, 20-25% of CRCs are familial, and only 5% are inherited forms either as familial adenomatous polyposis (FAP), Lynch syndrome, also known as hereditary non-polyposis colorectal cancer (HNPCRC), or MYH-gene associated polyposis (MAP) [2].
More importantly, local and systemic treatments by chemotherapeutic and targeting agents can increase survival rates in patients [3,4]; however, some side effects, along with resistance sound like deserve much more attention. Amongst targeting agents, the anti-vascular endothelial growth factor (VEGF)-VEGF receptor (VEGFR) targeted therapy has emerged for the ablation of tumor angiogenesis, preventing tumor proliferation and metastasis [5]. In this respect, Regorafenib (Stivarga®) as a multiple receptor tyrosine kinase inhibitor (RTKI) can block angiogenic, stromal, and oncogenic RTKs [6], and it has even received the Food and Drug Administration (FDA) approval as the last-line therapy for metastatic CRC (mCRC) and gastrointestinal stromal tumor (GIST) [7].
Although promising progress has been obtained regarding the treatment and overall survival of CRC patients, chemotherapy-induced drug-resistance is still a challenging phenomenon in this field [8]. It should be noted that various mechanisms, such as reduction of intracellular drug concentration, genetic or epigenetic alterations [8], apoptosis inhibition, tumor microenvironments, cancer stem cells (CSCs) [9], and epithelial-mesenchymal transition (EMT) [10], contribute to CRC drug-resistance. Moreover, mechanisms of resistance to anti-angiogenic therapies are attributed to substitute vasculogenesis systems, release of bone marrow-derived cells to tumor sites, local stromal cells [11], plethora of other angiogenic factors, and hypoxia [12]. It is believed that hypoxia is a key factor affecting resistance to Regorafenib [13] and other anti-angiogenic drugs, since it up-regulates the hypoxia-inducible factor 1-alpha (HIF-1α) and other downstream signaling pathways [14]. For example, HIF-1α induces up-regulation of Integrins, especially Integrin-β1 [12], EMT [11], and positive selection of CSCs [15].
Integrins are heterodimeric cell surface receptors [16], and they are able to transfer signals bi-directionally across transmembranes and participate in physiological and pathological processes. These receptors are also the drivers of stem cell-like, undifferentiated, mesenchymal phenotypes of cancer cells, inducing invasiveness, metastasis, as well as resistance in different types of cancer [17]. Integrin-β1 is also considered the greatest subset of Integrins, as it makes heterodimers with almost 2/3 of α subunits. Pre-clinical and clinical studies have further shown up-regulation and the prominent role of Integrin-β1 in the proliferation, survival, invasion, and metastatic features of tumor cells, and most importantly resistance to chemotherapy, radiotherapy, and anti-angiogenic targeted therapy [16,18].
Likewise, genetic therapy is currently developed to treat malfunctioned protein-based diseases by alteration of DNA and ribonucleic acid (RNA) as the progenitor of such proteins. RNA interference (RNAi) mechanism is also a specific and efficient gene silencing process targeting and degrading a specific RNA within the cytoplasm, thereby down-regulating corresponding genes [19]. Despite promising features of this novel treatment, several challenges have been raised about their systemic administration. The most important obstacles comprise degradation by serum endonucleases, rapid glomerular filtration by the kidneys, aggregation with serum proteins [20], up-taking by the reticuloendothelial system (RES), transmission through the vasculature, negatively charged lipid bilayer, stimulation of the innate immune system, off-target effects, and efficiency [21]. In this regard, various delivery systems have been introduced to address the aforementioned challenges. To deliver nucleic acids into cells, non-viral vectors like hybrid nanoparticles (HNPs) have received considerable attention compared to viral partners, since HNPs integrate the advantages of both polymeric and lipid-based nano-carriers [22] with the polymeric core to encapsulate hydrophobic drug and cationic lipid for nucleic acid loading [23]. Here, Dimethyldioctadecylammonium bromide (DDAB)-methoxy poly (ethylene glycol) (mPEG)-poly-ε-caprolactone (PCL) HNPs were used as biodegradable NPs with controlled release kinetics owing to the presence of PCL [24]. Furthermore, the PEG layer could enhance systemic circulation time and prevent it from renal filtration and uptaking by RES. The existence of DDAB as a cationic lipid on the shell could provide biocompatibility and facilitate siRNA delivery into cells [25].
To the best of our knowledge, the expression profile of Integrin-β1 has not been thus far evaluated in Regorafenib-resistant cell lines. Accordingly, Regorafenib-resistant human colon cancer cell line (SW-48) was established in this study for the first time. Then, siRNA-bearing HNPs, alone or in combination with HNPs encapsulating Regorafenib, were employed to assess such interventions on the Integrin-β1 expression to investigate the mechanisms involved in Regorafenib-resistant cell lines. The apoptosis rate in resistant cells was also evaluated using siRNA/HNPs, alone or in combination with Regorafenib/HNPs. It was assumed that Integrin-β1 targeting siRNA in combination with Regorafenib/HNPs had the potential effect to efficiently down-regulate this gene and to promote apoptosis, thereby overcoming drug resistance.
Materials and methods
Reagents and cell culture
The SW-48 cell line (ATCC: CCL-231) was obtained from the Pasteur Institute of Iran (Tehran, Iran). Regorafenib (sc-477163) was also purchased from Santa Cruz. Integrin-β1 siRNA (sense: 5’-UAGAUAUCUCGCGUCAUACdTdT-3’ and anti-sense: 5’-GUAUGACGCGAGAUAUCUAdTdT-3’) and control siRNA (sense: 5’-UAGAUAUCUCGCGUCAUACdTdT-3’ and anti-sense: 5’-GUAUGACGCGAGAUAUCUAdTdT-3’) were synthesized by Microsynth AG (Switzerland) and Eurofins Genomics (Germany), respectively. mPEG-PCL di-block co-polymer, which had been synthesized according to the ring-opening polymerization of caprolactone in the presence of mPEG, was received as a gift from Dr. Rostamizadeh [26]. Cationic lipid DDAB (D2779 10G), 4,5-dimethylthiazol-2-yl-2,5-diphenyltetrazolium bromide (MTT), and trypan blue were purchased from Sigma-Aldrich. Moreover, centrifugal filter Vivaspin Turbo 30 MWCO was obtained from Sartorius Stedim Biotech GmbH (Germany). Gel Red TM Nucleic Acid Gel Stain was acquired from Biotium (Fremont, United States). RPMI 1640 (GIBCO, Germany), trypsin EDTA (GIBCO, Germany), Penicillin-Streptomycin (Pen-Strep) (INNOCLON, Iran), fetal bovine serum (FBS) (BIOCHROM, Germany), dimethyl sulfoxide (DMSO) (Chem Cruz), SYBER green (AMPLIQON), along with all chemicals were prepared locally. Integrin-β1 and β-actin Primers were provided from SinaGene, Iran.
Resistant cell-line establishment
The sensitive cells were cultured in the RPMI 1640 medium supplemented with 10% FBS and 1% Pen-Strep in a humidified incubator at 37°C. The given cells were passaged once they reached confluency by 80%. To develop a resistant SW-48 cell line, the sensitive cells were maintained in a complete medium containing 40 μM Regorafenib for three days, and it was continued for five days without any drugs as recovery. This treatment-recovery cycle was repeated four times [27] (Table 1).
Table 1.
Cycles and duration of exposure to Regorafenib for establishment of drug-resistant SW-48 cell line
| Cycles | Cycle 1 | Cycle 2 | Cycle 3 | Cycle 4 |
|---|---|---|---|---|
| Treatment (Time of exposure/days) | 3 | 3 | 3 | 3 |
| Recovery (Time of drug-free/days) | 5 | 5 | 5 | 5 |
Half maximal inhibitory concentration (IC50) and MTT viability assay
To confirm resistance, IC50 of Regorafenib was measured in a non-resistant and resistant SW-48 cell line using the MTT assay. For this purpose, both groups of cells were seeded in a 96-well plate at a density of 6000 cells/well in triplicate, containing 100 μl of the complete medium supplemented with 10% FBS. After incubation for 24-48 hours at 37°C in the presence of 5% carbon dioxide (CO2), the cells were washed with phosphate-buffered saline (PBS) and exposed to different doses of Regorafenib in the RPMI medium for 12 hours. Then, the supernatant was aspirated and replaced with the complete medium. After 24-hour treatment, 20 μl of the MTT solution (5 mg/ml in PBS) was added following incubation for 3-4 hours. Once the liquid was discarded, 200 μl of DMSO was added to each well to dissolve the formazan crystals. Finally, optical density (OD) was measured at 570 nm, using a plate reader. Untreated cells were considered a control group. The percentage of cell relative viability was calculated as the following formula:
Cell relative viability = Mean OD of treated cells/mean OD of untreated cells ×100.
The viability percentage was also plotted against the Regorafenib concentration in each group.
Likewise, the MTT assay was performed to evaluate the viability of the cells at the end of each cycle and after resistance. Briefly, the frozen cells at the end of the cycles were revived and seeded in a 96-well plate with 6000 cells per well in triplicate. After incubation for 24 hours at 37°C in the presence of 5% CO2, the cells were washed with PBS and exposed to the RPMI medium containing 40 μM Regorafenib, for 24 hours. The MTT assay was adjusted to the protocol described in the previous section.
Preparation of DDAB-mPEG-PCL HNPs
DDAB-mPEG-PCL HNPs were synthesized using the single-step nano-precipitation method, according to the protocol reported in the previous study [26]. Briefly, 800 μl of the co-polymer stock solution (2.5 mg/ml in acetonitrile) were mixed with 200 μl of the DDAB stock solution (1 mg/ml in distilled water). Then, 7800 μl of distilled water was rapidly injected into the solution by syringes (5 ml), and the mixture was further sonicated for 6 minutes at 50°C. Synthesized HNPs were concentrated in Vivaspin filter (molecular weight cutoff of 30 KDa) through centrifugation, at 5000 rpm for 30 minutes to the final volume of 500 μl.
Preparation of Regorafenib/DDAB-mPEG-PCL HNPs
To synthesize Regorafenib/DDAB-mPEG-PCL HNPs, 800 μl of the co-polymer containing Regorafenib stock solution (2.5 mg/ml co-polymer with 0.0625 mg/ml Regorafenib in acetonitrile and DMSO as solvents) and 200 μl of DDAB stock solution (1 mg/ml in distilled water) were mixed together, and then, 7800 μl of distilled water was rapidly injected into the solution by syringes (5 ml). Subsequently, the mixture was sonicated for 6 minutes at 50°C. The concentration was carried out by Vivaspin columns.
Characterization of synthesized HNPs
Malvern Zeta sizer Nano ZS90 apparatus (Malvern Instruments, Worcestershire, United Kingdom) was used to specify the size, the surface charge, and the polydispersity index (PDI) of HNPs. However, for dilution purposes, the OD of HNPs was measured in 633 nm, using a spectrophotometer.
Preparation of siRNA-loaded HNPs and gel retardation assay
To fulfill efficient delivery, certain amounts of the HNPs were combined with a fixed concentration of siRNA (52 ng) to form siRNA/HNP complexes with different ratios of positively charged polymer amine nitrogen (N) to negatively charged nucleic acid phosphate (P) (N/P ratio). The volume of the HNPs employed was as follows:
Volume of HNPs (μl) = μg of siRNA × nmol of phosphate per 1 μg × N/P ratio/nmol of nitrogen in cationic lipid per 1 μg.
It should be noted that there are 3 nmol of phosphate in one microgram of siRNA and 0.634 nmol of nitrogen in one microgram of DDAB as a cationic lipid. Therefore, various N/P ratios need different volumes of HNPs.
For HNP/siRNA complex formation, the mixtures were incubated for 1 hour at room temperature. SiRNA/HNP complexes were then run on a 2% agarose gel for gel retardation assay at a constant voltage of 120 V for 30-45 minutes in tris-borate-EDTA (TBE) 1× buffer and stained with Gel Red dye developed using UV- Transilluminator (Analitic Jena AG). Free siRNA was also used as a positive control.
Determination of siRNA encapsulation efficiency
Optimized volumes of HNPs were mixed with siRNA and incubated for 1 hour at room temperature to form the complex, which was then centrifuged at 5000 rpm for 5 minutes. The absorbance of the total amount of siRNA and free siRNA, which had been already passed through the filter, was also measured by a Nanodrop at 260 nm to evaluate the encapsulation efficiency. The following formula was utilized for the encapsulation efficiency (EE) calculation:
EE = (total amount of siRNA-free siRNA in underlaid solution)/total amount of siRNA ×100.
Treatments and siRNA delivery by HNPs
To evaluate the effects of siRNA/HNPs and Regorafenib/HNPs on the Integrin-β1 gene silencing and apoptosis of cancer cells, 3.5×105 cells were seeded into 6-well plates, containing 2 ml of enriched media, and incubated for 48 hours at 37°C. Then, different concentrations of siRNA (25, 50, and 100 nM) and Regorafenib (40 µM), their HNP complexes, as well as their combinations (Table 2) were transfected into the cells in FBS/antibiotic-free RPMI. After 16-hour (overnight) incubation, the transfection medium was replaced with the enriched one and further incubated. RNA isolation, complementary DNA (cDNA) synthesis, and gene expression studies were conducted after 36 hours, and the apoptosis assay was assessed after 56 hours.
Table 2.
Treatment groups of resistant SW-48 cells and different doses of siRNA, siRNA/HNPS, free drug, and drug-loaded HNPs
| Group | Sub-group |
|---|---|
| A) SiRNA | A1: siRNA 100 |
| A2: SiRNA 50 | |
| A3: SiRNA 25 | |
| B) SiRNA/HNP | B1: SiRNA/HNP 100 |
| B2: SiRNA/HNP 50 | |
| B3: SiRNA/HNP 25 | |
| C) Drug40+siRNA/HNP | C1: Drug 40+SiRNA/HNP 100 |
| C2: Drug 40+SiRNA/HNP 50 | |
| C3: Drug 40+SiRNA/HNP 25 | |
| D) NPD40+siRNA/HNP | D1: NPD40+siRNA/HNP 100 |
| D2: NPD40+siRNA/HNP 50 | |
| E) others | E1: SiRNA control/HNP 100 |
| E2: NP100 | |
| E3: Drug 40 | |
| E4: NPD40 | |
| E5: NPD20 | |
| F) control | F: untreated resistant cells |
RNA isolation and cDNA synthesis
Total RNA from the transfected and control cells were extracted using the TrizoLEX reagent (DNAbiotech), as stated in the manufacturer’s instructions. The concentration of the extracted RNAs, as well as the 260/280 and 260/230 ratios, was determined to identify purity and contamination with chloroform and isopropanol using Nanodrop (Thermo). In addition, the integrity of RNAs was qualified by employing 2% agarose gel electrophoresis. The cDNA synthesis kit (Yekta Tajhiz Azma, Iran (YT4500)) was applied to convert the extracted RNAs into cDNA. Furthermore, 5 μg of RNA was correspondingly used as an RNA template for each sample, and it reached the final volume of 19.9 μl by adding other ingredients of the kit.
Real-time polymerase chain reaction (RT-PCR)
For this purpose, 1 μl of synthesized cDNA was combined with 0.5 μl of each Forward/Reverse primer as well as deionized water and RealQ Plus Master Mix Green (AMPLIQON). The PCR reaction was conducted using the ABI (StepOnePlus) system. The sequences of the Integrin-β1 primers were Forward: 5’ TGATTGGCTGGAGGAATGTTA 3’ and Reverse: 5’ GTTTCTGGACAAGGTGAGCAA 3’. β-actin was additionally recruited as the housekeeping control gene, and the primer sequences were Forward: 5’ ACTCTTCCAGCCTTCCTTCC 3’ and Reverse: 5’ CGTACAGGTCTTTGCGGATG 3’. PCR production length was 186 bp for Integrin-β1 and 104 bp for β-actin. The PCR cycling condition consisted of an initial activation step at 95°C for 15 minutes and 40 cycles of 95°C for 15 seconds (sec), 60°C for 30 sec, and 72°C for 30 sec. The relative expression level of Integrin-β1 was further normalized to β-actin and untreated control group levels. Ultimately, the fold change was calculated using the E-ΔΔCT formula to represent the data. To ensure the quality of the products, RT-PCR aliquots were run on 2% agarose gel electrophoresis and then visualized using an UV set after being dye-stained.
Apoptosis assay and flow cytometry (FC)
The transfection of the resistant cells was performed as just described for the Integrin-β1 expression assay using RT-PCR. Cell apoptosis assay was conducted according to the protocol of the FITC Annexin V Apoptosis Detection Kit (Invitrogen/eBioscience), 56 hours after transfection. Briefly, the cells were trypsinized and washed with cold PBS and 1× binding buffer. Subsequently, the cell suspension in the 1× binding buffer was prepared at the concentration of 1-5×106/ml. Then, 2.5 μl of FITC Annexin V was added to 100 μl of the cell suspension and further incubated for 10-15 minutes at RT. After being washed with 1× binding buffer, the cells were resuspended in 200 μl binding buffer, and 2.5 μl of propidium iodide (PI) was added to the samples. The rate of apoptosis was assessed using the BD FACSCalibur instrument. Both early apoptotic (Annexin V-positive and PI-negative) and late apoptotic (Annexin V-positive and PI-positive) cells were considered total apoptosis determinants. The results were analyzed by the FlowJO software. However, the flow cytometer was calibrated using instrument-related fluorescence beads to obtain accurate data. The control cells (namely, unstained and stained) were also employed to adjust the fluorescence threshold.
Statistical analysis
Statistical analysis was performed using the IBM SPSS Statistics software (version 24), and the related graphs were drawn by GraphPad Prism 8.0.1. Kolmogorov-Smirnov and Shapiro-Wilk tests were conducted to assess normal distribution. To compare the differences between the resistant and non-resistant variables, an independent-samples t-test was performed. One-way analysis of variance (ANOVA) with Tukey’s post-hoc test was also conducted to compare the differences between multiple treatments. All the data were expressed as the mean ± standard deviation (SD), and p-value <0.05 was considered statistically significant.
Results
Establishment of regorafenib-resistant SW-48 cell line
To confirm the development of resistant SW-48 cells, the MTT assay was performed. As Figure 1 shows, the cell viability percentage at the end of the eighth day was 73.8±7.9% in cycle 1, 86.8±3.1% in cycle 2, 90.2±0.9% in cycle 3, and 92.3±0.8% in the last cycle. Cell viability in all of the cycles was significantly decreased after the Regorafenib treatment compared to the control cells. Once the cycles were analyzed, a significant increase in cell viability was observed from the first cycle toward the last one, indicating the adaptation of the cells to the drug. In addition, the IC50 of Regorafenib was measured in resistant and non-resistant cells, using the MTT method. As Figures 2 and 3 display, the IC50 was 13.5±1.5 μM for non-resistant cells, but it augmented to 55.1±0.84 μM in resistant ones, indicating that the cells could tolerate the drug owing to resistance and adaptation.
Figure 1.
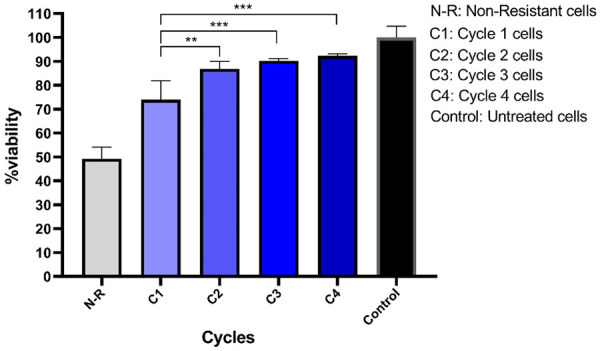
Cell survival percentage in the SW-48 cell line at the end of each cycle using the MTT method. The cells were treated for four cycles by Regorafenib, and then the MTT assay was conducted. They experienced a high rate of mortality in the early cycles, but adapted to the environment and exhibited more viability at the end of cycles 3 and 4. According to the Tukey’s post-hoc test of multiple comparisons, significant differences were observed between each cycle, cycle 1 and the non-resistant group. **P=0.0017, ***P<0.0001.
Figure 2.
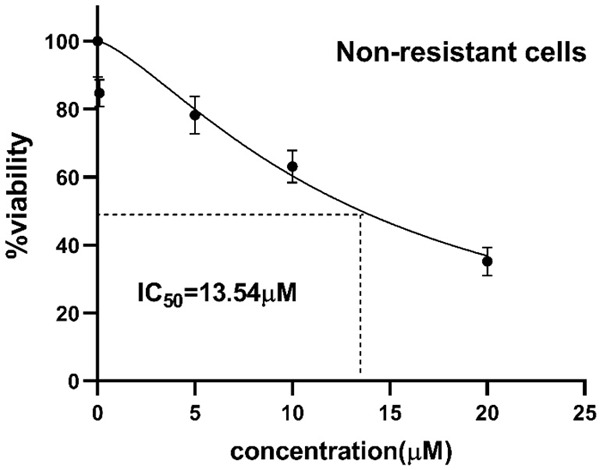
Inhibitory effect of Regorafenib on the cell viability of non-resistant SW-48 cells in a dose-dependent manner. Cell viability was measured using the MTT method by incubating the cells to the increasing doses of Regorafenib (0-20 μM) for 24 hours. The IC50 was 13.5±1.5 μM in non-resistant cells.
Figure 3.
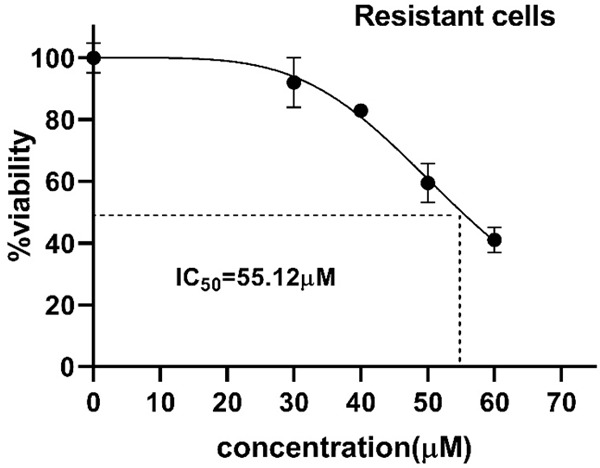
Inhibitory effect of Regorafenib on the cell viability of resistant SW-48 cells in a dose-dependent manner. Cell viability was measured using the MTT method by incubating the cells to the increasing doses of Regorafenib (0-60 μM) for 24 hours. The IC50 was 55.1±0.84 μM in resistant cells.
DDAB-mPEG-PCL HNPs and regorafenib/HNPs characterization
The DDAB-mPEG-PCL HNPs and Regorafenib/HNPs were prepared using the single-step nano-precipitation method. Dynamic light scattering (DLS) was further used to control the quality of the HNPs. As Table 3, and Figure 4A and 4B show, Z-average size, zeta potential, and PDI of DDAB-mPEG-PCL HNPs were 66.56±0.5 nm, +29.5±1.2 mv, and 0.327±0.08, respectively. As Figure 5A and 5B illustrate, Z-average size, zeta potential, and PDI of Regorafenib-loaded HNPs were also equal to 68.25±1.1 nm, +26.9±0.6 mv, and 0.371±0.04, respectively.
Table 3.
Size distribution, zeta potential, and PDI of DDAB-mPEG-PCL HNPs and Regorafenib loaded HNPs
| NP | Size (nm) ± SD | Zeta potential (mV) ± SD | PDI ± SD |
|---|---|---|---|
| DDAB-mPEG-PCL HNPs | 66.56±0.5 | +29.5±1.2 | 0.327±0.08 |
| Regorafenib-loaded HNPs | 68.25±1.1 | +26.9±0.6 | 0.371±0.04 |
Figure 4.
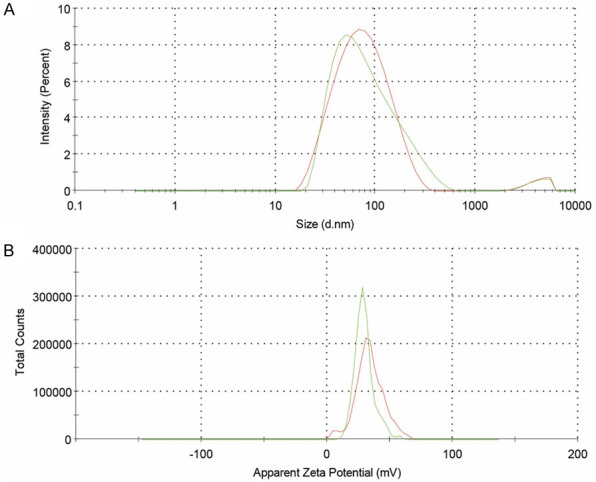
Size distribution (A) and zeta potential (B) of DDAB-mPEG-PCL HNPs.
Figure 5.
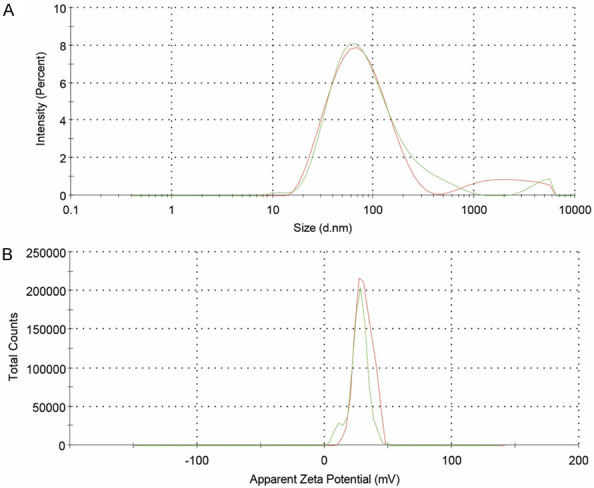
Size distribution (A) and zeta potential (B) of Regorafenib loaded HNPs.
SiRNA/HNP complex formation
Gel retardation assay was conducted to confirm the formation of siRNA/HNP complexes and to determine the optimum N/P ratio for maximum efficacy. Therefore, HNPs and siRNA were combined at the ratio of 0-60 to form the complexes. After being incubated for 1 hour, the samples were run on 2% agarose gel. The wells related to the N/P ratios of 2-15 exhibited obvious bands similar to the control one, while there was no band on the well related to the N/P ratio of 20. Thus, the N/P ratio of 20 was selected as the best N/P ratio, since higher N/P ratios than 20 result in more cytotoxicity to the cells (Figure 6).
Figure 6.

N/P ratios of 0-60 on the agarose gel to determine the optimum N/P ratio for maximum binding of siRNA on HNPs’ surface. The N/P ratio of 20 was selected as the best N/P ratio.
SiRNA encapsulation efficiency of HNPs
Ultrafiltration of the siRNA/HNP complexes at the N/P ratio of 20 was obtained using gel retardation assay. Then, the absorbance of the total amount of siRNA added to the HNP and free siRNA passed through the Vivaspin column was measured at 260 nm. siRNA encapsulation efficiency was further calculated to be 98.7% at the N/P ratio of 20. Hence, the majority of siRNA molecules were tightly attached to the NP via different interactions, especially electrostatic bond ones.
Integrin-β1 expression in resistant and non-resistant SW-48 cell lines
The Integrin-β1 gene expression level was measured in resistant cells using RT-PCR and then compared to non-resistance. The analysis of the obtained cycle threshold (CTs) showed a significant difference in the Integrin-β1 mRNA level between resistant and non-resistant cells (P<0.05). In this regard, the changes in the mRNA folds in resistant cells were 1.81 times as much as non-resistant ones (Figure 7), confirming the Integrin involvement in cancer resistance.
Figure 7.
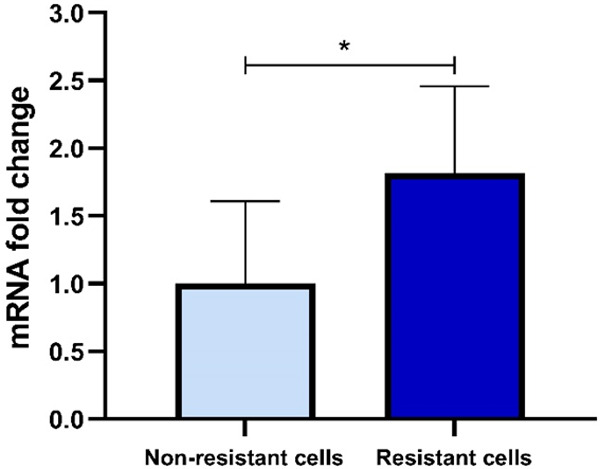
Integrin-β1 gene expression levels in resistant and non-resistant SW-48 cell lines. Integrin-β1 mRNA fold change showed approximately 1.8 times higher value than that of the non-resistant one, which was statistically significant (P<0.05).
Down-regulation of integrin-β1 using a combination of sirna/hnps and regorafenib/hnps in resistant sw-48 cells
Statistically, the data analysis demonstrated that the use of free siRNA, HNPs, and control siRNA did not influence the Integrin-β1 gene expression. However, B1 (siRNA-HNP with 100 nM of siRNA) and B2 (namely, siRNA-HNP with 50 nM of siRNA) subgroups (in the B group) significantly diminished the Integrin-β1 mRNA level compared to the control group (P<0.01), demonstrating the importance of HNPs in siRNA delivery. Combination therapy with D40 (Regorafenib 40 µM) and siRNA/HNPs as well as NPD40 (HNP bearing 40 µM of Regorafenib) and siRNA/HNPs showed different results. Accordingly, the C1 subgroup exhibited the most significant effects on the Integrin-β1 mRNA level down-regulation (P<0.001). Furthermore, the combination of NPD40 and siRNA/HNPs either by D1 or D2 sub-groups significantly mitigated the mRNA level (P<0.001). The NPD40 with siRNA/HNP 100 was also found as the most effective treatment in decreasing the Integrin-β1 gene expression. In other words, Regorafenib-loaded HNPs, along with siRNA/HNPs had the highest capacity to down-regulate the Integrin-β1 gene. Overall, HNPs facilitated siRNA and drug delivery into cancer cells, and the combination of both targeting agents exhibited synergistic effects on the cells (Figures 8 and 9).
Figure 8.
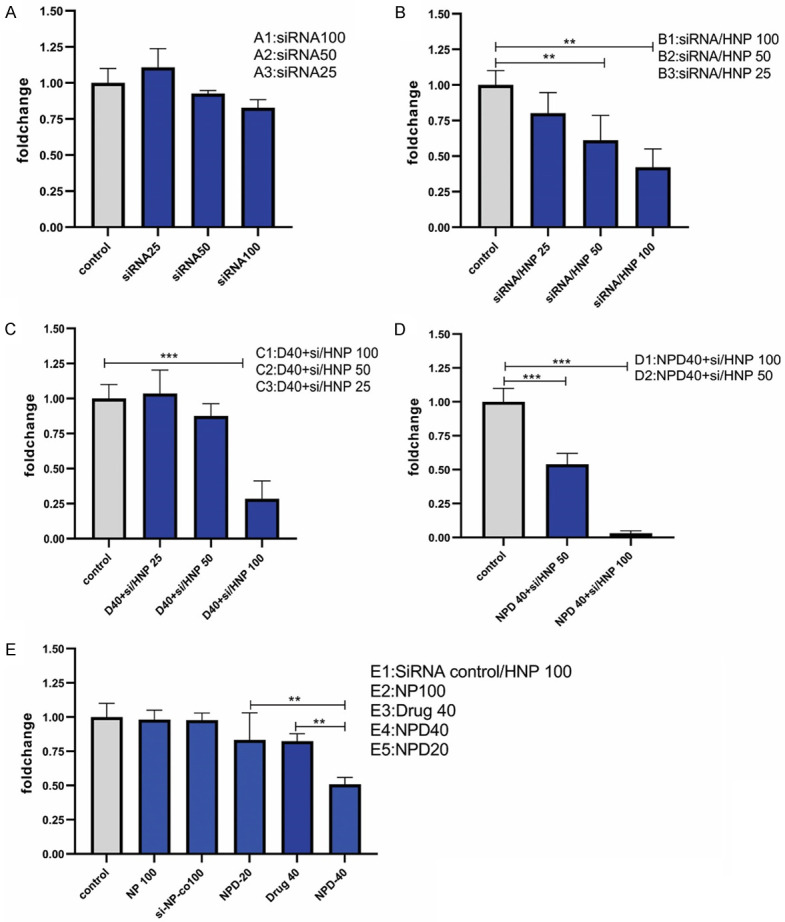
Comparison of the Integrin-β1 gene expression level in different treatment groups of resistant SW-48 cells. The use of free siRNA did not significantly affect Integrin-β1 gene expression (A). Significant differences were observed in the subgroups of (B-D) compared to the untreated control group. (E) Comparison of the effect of Regorafenib-loaded HNPs and free Regorafenib on the Integrin-β1 gene expression. HNPs and siRNA negative control/HNPs were used as controls. **P<0.01, ***P<0.001.
Figure 9.
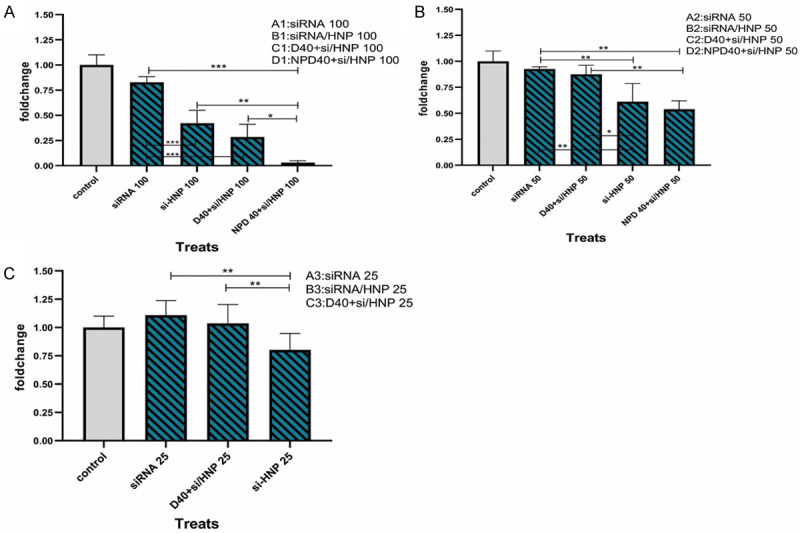
Intragroup comparison of the Integrin-β1 gene expression in resistant SW-48 cells. Comparison of the effect of 100 μΜ (A), 50 μΜ (B) and 25 μΜ (C) doses of siRNA when using alone, siRNA/HNP complexes, combination of D40 and siRNA/HNPs, as well as NPD40 and siRNA/HNPs. *P<0.05, **P<0.01, ***P<0.001.
Apoptosis effects of siRNA/HNPs targeting integrin and regorafenib/HNPs in resistant SW-48 cells
The FC results showed that using siRNA alone did not influence the apoptosis rate, while siRNA/HNP 100 nM could enhance it by 22.27±0.61%. The combination therapy of D40 with siRNA/HNP 100 nM also induced apoptosis by 37±1.6%. In this respect, NPD40 with siRNA/HNP 100 nM had the most significant effects on the apoptosis of resistant cells by 85.4±1.7% (P<0.001). Simply put, Regorafenib could trigger the apoptosis signaling in the cell through down-regulation of the Integrin-β1 gene by siRNA/HNPs (Figures 10 and 11).
Figure 10.
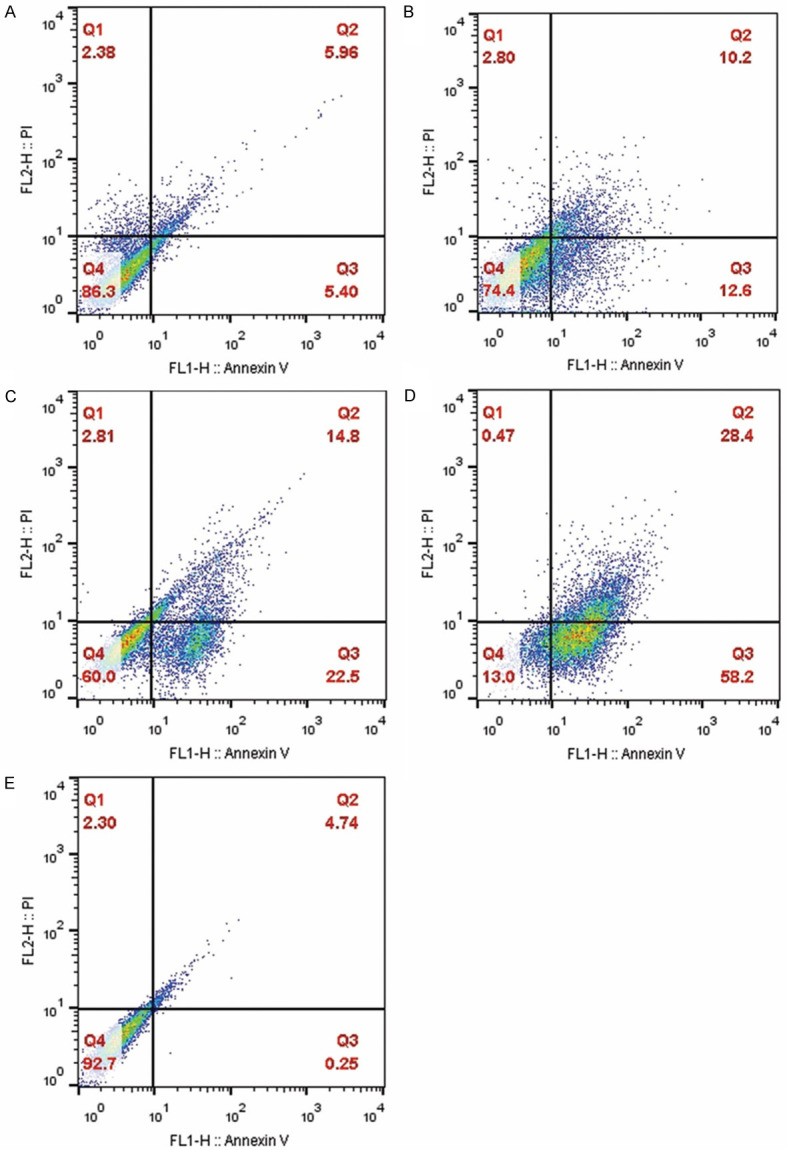
Effect of Integrin-β1 siRNA 100 (A), siRNA-HNP 100 (B), D40+siRNA-HNP 100 (C), and NPD40+siRNA-HNP 100 (D) on the apoptosis rate of resistant SW-48 cells detected by FC. Q1, Q2, Q3, and Q4 quadrants represent necrotic, early apoptotic, late apoptotic, and normal cells, respectively. (E) show the control group (i.e., untreated group).
Figure 11.
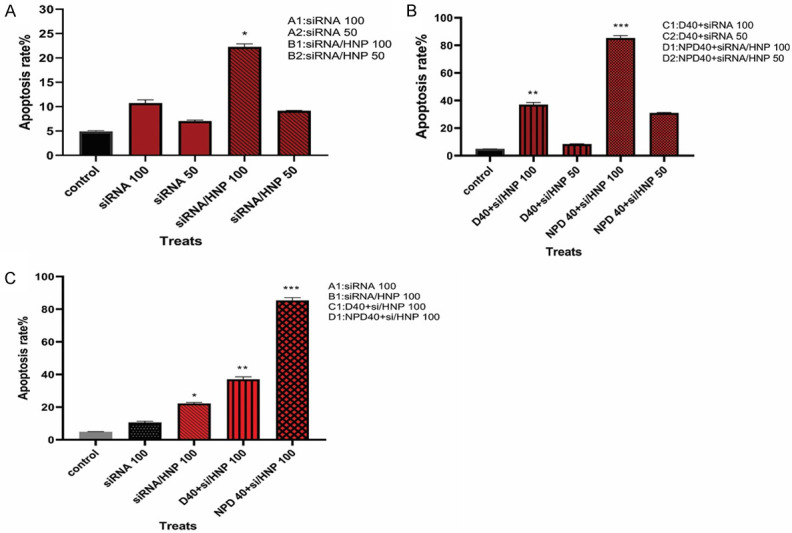
Comparison of the apoptosis rate in different treatment groups. The apoptosis assay was conducted using FC after being treated with different formulations of Integrin-β1 siRNA. Both early apoptotic (Annexin V+/PI-) and late apoptotic (Annexin V+/PI+) cells were considered the determinants of cell death.
Discussion
Drug resistance to anti-angiogenic therapies like Regorafenib deserves further attention due to its effects on survival rates in metastatic patients. Accordingly, studies have revealed that one of the main mechanisms related to resistance to these agents correlates with hypoxia and HIF-1α up-regulation [28]. In this respect, HIF-1α activates the cMET-HGF signaling pathway, up-regulates the Integrin-β1 gene expression, and induces malignant phenotypes in cancer [12]. Therefore, the Integrin-β1 gene silencing by siRNA can be an effective and specific method to combat such resistance. Here, the Regorafenib-resistant SW-48 cell line mimicking in vivo cancer resistance was established according to previous experiments. This report demonstrated that anti-Integrin-β1 siRNA using DDAB-mPEG-PCL HNPs as a carrier had the potential to down-regulate the Integrin-β1 gene, resulting in releasing the apoptosis pathway that could be induced via combination with Regorafenib/HNPs.
In the previous study, a gradual method was used to develop the resistant SW-48 cell line considering that resistance could enrich cancer stem cells [29]. In this study, resistant SW-48 cells were induced by exposing them to Regorafenib using the cyclic method [27] to describe that cancer cells could tolerate chemotherapeutic agents, and the mode of exposure was unimportant. The IC50 of Regorafenib was additionally increased in resistant cells compared to non-resistant ones, and the viability of the cells was enhanced with the time of receiving the drug, implying the adaptation of the cells to the drug. Although IC50 has indicated different values in various studies on investigated cancer resistant cells, it has usually been higher in resistant cancer cells than that in non-resistant ones. Given that cancer cells have genome instability and heterogeneity, these differences in IC50 among various studies may be attributed to various cell lines, resistance induction protocols, passage number of cells, drug concentrations, origins of cells, and so forth.
The DDAB-mPEG-PCL HNPs and Regorafenib/HNPs were synthesized in this study using the single-step nano-precipitation method, as described by Fang [30]. The Z-average size of the prepared DDAB-mPEG-PCL HNPs was 66.56±0.5 nm, which is appropriate for the systemic delivery of siRNA to the tumor site. Moreover, these HNPs had the zeta potential of +29.5±1.2 mv for effective encapsulation of siRNA and cellular uptake.
The N/P ratio of the siRNA/DDAB-mPEG-PCL complexes was 20. Lu Y investigated 1,2-dioleoyloxy-3-(trimethylammonium) propane (DOTAP)-conjugated mPEG-PCL (DMP) NP to bring myeloid cell leukemia 1 (Mcl-1) and B-cell lymphoma-extra-large (Bcl-xL) siRNA into CRC cells. The size of this nano-micelle was 144.8 nm with the zeta potential of +46.4 mv and the N/P ratio equal to 30. Thus, it was concluded that siRNA-loaded DMP micelles could decrease gene expression and trigger apoptosis, thereby inhibiting tumor growth in C26 cells and in vivo xenograft models [31].
Yang used N,N-bis(2-hydroxyethyl)-N-methyl-N-(2-cholesteryloxycarbonylaminoethyl) ammoniumbromide (BHEM-Chol)/mPEG-PLA HNPs to deliver polo-like kinase 1 (PLK-1) specific siRNA into the human breast cancer (BT-474) cell line with the particle size of 110 nm, the zeta potential of +50 mV (in lipid/polymer weight ratio of 0.1), and the N/P ratio of 5. These HNPs had high encapsulation efficiency up to 90% and were also able to down-regulate the PLK-1 gene considerably, induce apoptosis, and decline tumor growth in vitro and in vivo [32].
The N/P ratio describes the relative numbers of positive charges, present on the NP for efficient binding with the negatively charged cell membrane. Thus, it could play a pivotal role in the cell uptaking of the cargo. It appears that the size and the zeta-potential can directly affect the N/P ratio. Furthermore, in a previous study conducted to investigate the delivery efficiency of PLA-PEG-PLA tri-block copolymer and DDAB, the N/P ratio was obtained in 60 [26]. Accordingly, different structures in the polymer and cationic lipid may play a crucial role in the distribution and organization of charges and ultimately the N/P ratio.
In addition, activation of Integrin-β1 downstream signaling pathways results in apoptosis resistance, survival, proliferation, invasion, and metastasis of cancer cells [16,33]. Tumor microenvironment, cancer stem cells [34], and EMT [10] are the major contributors to the Integrin-β1 function to induce cancer drug resistance. Different studies have also demonstrated the up-regulation of Integrin-β1 in response to resistance to radiotherapy, chemotherapy, targeted therapy, and anti-angiogenic drugs [33,35,36]. For example, Michael DeLay found the Integrin-β1 4-fold overexpression in the clinical samples of bevacizumab-resistant patients, and similarly Carbonell demonstrated the 8-fold up-regulation of this protein in Bevacizumab-treated BRG cells [28,37]. However, there is no study evaluating the Integrin-β1 expression in Regorafenib-resistant SW-48 cell lines. The results of the present study indicated that the Integrin-β1 gene expression could increase by 1.81 times in resistant cells than in non-resistant ones.
Integrin-β1 as the largest subgroup of Integrins [16] may be compared to the β subunits of human chorionic gonadotropin (hCG), which is a dominant contributor in the tumors derived from embryonic stem cells. Integrin-β1 is possibly a major player in the deviation of cancer stem cells from original tumor ones and may be introduced as a therapeutic target to disrupt the hallmarks of resistant cancer. In line with these experiments, Mu investigated the effect of anti-c-Jun N-terminal kinase-1 (Jnk-1) siRNA using 1,2-Distearoyl-sn-glycero-3-phosphorylethanolamine (DSPE)/PEG-PEI-PLGA NPs in the human prostate cancer (DU145) cell line, and concluded that it could induce the apoptosis; however, no combination therapy with chemotherapeutic agents was utilized [38]. To achieve the maximum effect of Integrin-β1 siRNA, different concentrations were tested, confirming that the combination of siRNA/HNPs with Regorafenib/HNPs was the most effective treatment in decreasing the Integrin-β1 gene expression and the apoptosis induction in resistant cells. The results of this study indicated that the Integrin-β1 mRNA level was significantly decreased, when the resistant cells were treated with the combination of siRNA/HNP and Regorafenib/HNPs. The results demonstrated that the release of Integrin-β1-mediated anti-apoptosis could be challenged by specific siRNA, which could finally lead to the effectiveness of Regorafenib. Dong indicated that down-regulation of survivin in combination with Paclitaxel and Epirubicin in the resistant breast cancer cell line (MCF-7) could trigger apoptosis, decrease cell proliferation, and reverse the resistance process [39].
Resistance to targeted therapy through Integrin-β1 in solid tumors occurs via different mechanisms. In addition, Integrin-β1 can employ RTKs or other co-receptors to develop such resistance. Despite the prominent therapeutic role of Integrin-β1, the complicated and heterodimeric structure of these glycoproteins as well as the role of α sub-units should be considered. Using Integrin-β1 as a therapeutic target in combination with chemotherapeutic drugs is also a promising method to classify patients, reduce dosage of therapeutic agents, enhance the effect of chemotherapy, and minimize side effects.
Conclusion
Induction of resistant SW-48 CRC cell lines attenuated the anti-cancer effects of Regorafenib and up-regulated the Integrin-β1 gene expression in these cells. The study also signified the role of combination therapy using siRNA/HNPs with Regorafenib/HNPs to down-regulate the Integrin-β1 gene expression and induce apoptosis remarkably in resistant cells, which might influence the Regorafenib-resistant signaling pathway.
Acknowledgements
This study was funded by grants (A-12-802-23 and A-12-802-27) from Zanjan University of Medical Sciences (ZUMS), School of Medicine, Cancer Gene Therapy Research Center, and Department of Clinical Biochemistry.
Disclosure of conflict of interest
None.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Binefa G, Rodríguez-Moranta F, Teule À, Medina-Hayas M. Colorectal cancer: from prevention to personalized medicine. World J Gastroenterol. 2014;20:6786–808. doi: 10.3748/wjg.v20.i22.6786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hagan S, Orr MC, Doyle B. Targeted therapies in colorectal cancer-an integrative view by PPPM. EPMA J. 2013;4:3. doi: 10.1186/1878-5085-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mody K, Baldeo C, Bekaii-Saab T. Antiangiogenic therapy in colorectal cancer. Cancer J. 2018;24:165–170. doi: 10.1097/PPO.0000000000000328. [DOI] [PubMed] [Google Scholar]
- 5.Vasudev NS, Reynolds AR. Anti-angiogenic therapy for cancer: current progress, unresolved questions and future directions. Angiogenesis. 2014;17:471–494. doi: 10.1007/s10456-014-9420-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Skårderud MR, Polk A, Vistisen KK, Larsen FO, Nielsen DL. Efficacy and safety of Regorafenib in the treatment of metastatic colorectal cancer: a systematic review. Cancer Treat Rev. 2018;62:61–73. doi: 10.1016/j.ctrv.2017.10.011. [DOI] [PubMed] [Google Scholar]
- 7.Rey JB, Launay-Vacher V, Tournigand C. Regorafenib as a single-agent in the treatment of patients with gastrointestinal tumors: an overview for pharmacists. Target Oncol. 2014;33:189–205. doi: 10.1007/s11523-014-0333-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hammond WA, Swaika A, Mody K. Pharmacologic resistance in colorectal cancer: a review. Ther Adv Med Oncol. 2016;8:57–84. doi: 10.1177/1758834015614530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mansoori B, Mohammadi A, Davudian S, Shirjang S, Baradaran B. The different mechanisms of cancer drug resistance: a brief review. Adv Pharm Bull. 2017;7:339–348. doi: 10.15171/apb.2017.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Du B, Shim JS. Targeting epithelial-mesenchymal transition (EMT) to overcome drug resistance in cancer. Molecules. 2016;21:965. doi: 10.3390/molecules21070965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Beijnum JR, Nowak-Sliwinska P, Huijbers EJ, Thijssen VL, Griffioen AW. The great escape; the hallmarks of resistance to antiangiogenic therapy. Pharmacol Rev. 2015;67:441–461. doi: 10.1124/pr.114.010215. [DOI] [PubMed] [Google Scholar]
- 12.Itatani Y, Kawada K, Yamamoto T, Sakai Y. Resistance to anti-angiogenic therapy in cancer-alterations to anti-VEGF pathway. Int J Mol Sci. 2018;19:1232. doi: 10.3390/ijms19041232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tomida C, Nagano H, Yamagishi N, Uchida T, Ohno A, Hirasaka K, Nikawa T, Teshima-Kondo S. Regorafenib induces adaptive resistance of colorectal cancer cells via inhibition of vascular endothelial growth factor receptor. J Med Invest. 2017;64:262–265. doi: 10.2152/jmi.64.262. [DOI] [PubMed] [Google Scholar]
- 14.Mansour RN, Enderami SE, Ardeshirylajimi A, Fooladsaz K, Fathi M, Ganji SM. Evaluation of hypoxia inducible factor-1 alpha gene expression in colorectal cancer stages of Iranian patients. J Cancer Res Ther. 2016;12:1313–1317. doi: 10.4103/0973-1482.199542. [DOI] [PubMed] [Google Scholar]
- 15.Vinogradov S, Wei X. Cancer stem cells and drug resistance: the potential of nanomedicine. Nanomedicine. 2012;7:597–615. doi: 10.2217/nnm.12.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cruz da Silva E, Dontenwill M, Choulier L, Lehmann M. Role of Integrins in resistance to therapies targeting growth factor receptors in cancer. Cancers. 2019;11:692. doi: 10.3390/cancers11050692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seguin L, Desgrosellier JS, Weis SM, Cheresh DA. Integrins and cancer: regulators of cancer stemness, metastasis, and drug resistance. Trends Cell Biol. 2015;25:234–240. doi: 10.1016/j.tcb.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pouliot N, Nice EC, Burgess AW. Laminin-10 mediates basal and EGF-stimulated motility of human colon carcinoma cells via α3β1 and α6β4 Integrins. Exp Cell Res. 2001;266:1–10. doi: 10.1006/excr.2001.5197. [DOI] [PubMed] [Google Scholar]
- 19.Rossor AM, Reilly MM, Sleigh JN. Antisense oligonucleotides and other genetic therapies made simple. Pract Neurol. 2018;18:126–131. doi: 10.1136/practneurol-2017-001764. [DOI] [PubMed] [Google Scholar]
- 20.Tatiparti K, Sau S, Kashaw SK, Iyer AK. siRNA delivery strategies: a comprehensive review of recent developments. Nanomaterials. 2017;7:77. doi: 10.3390/nano7040077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karim M, Tha KK, Othman I, Borhan Uddin M, Chowdhury EH. Therapeutic potency of nanoformulations of siRNAs and shRNAs in animal models of cancers. Pharmaceutics. 2018;10:65. doi: 10.3390/pharmaceutics10020065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mukherjee A, Waters AK, Kalyan P, Achrol AS, Kesari S, Yenugonda VM. Lipid-polymer hybrid nanoparticles as a next-generation drug delivery platform: state of the art, emerging technologies, and perspectives. Int J Nanomedicine. 2019;14:1937–1952. doi: 10.2147/IJN.S198353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wakaskar RR. General overview of lipid-polymer hybrid nanoparticles, dendrimers, micelles, liposomes, spongosomes and cubosomes. J Drug Target. 2018;26:311–318. doi: 10.1080/1061186X.2017.1367006. [DOI] [PubMed] [Google Scholar]
- 24.Ramanujam R, Sundaram B, Janarthanan G, Devendran E, Venkadasalam M, Milton MJ. Biodegradable polycaprolactone nanoparticles based drug delivery systems: a short review. Biosci Biotechnol Res Asia. 2018;15:679–685. [Google Scholar]
- 25.Madni A, Tahir N, Rehman M, Raza A, Mahmood MA, Khan MI, Kashif PM. Hybrid Nano-carriers for potential drug delivery. Adv Technol Deliver Ther. 2017:53–87. [Google Scholar]
- 26.Monirinasab H, Asadi H, Rostamizadeh K, Esmaeilzadeh A, Khodaei M, Fathi M. Novel lipid-polymer hybrid nanoparticles for siRNA delivery and IGF-1R gene silencing in breast cancer cells. J Drug Deliv Sci Technol. 2018;48:96–105. [Google Scholar]
- 27.Tong J, Tan S, Zou F, Yu J, Zhang L. FBW7 mutations mediate resistance of colorectal cancer to targeted therapies by blocking Mcl-1 degradation. Oncogene. 2017;36:787–796. doi: 10.1038/onc.2016.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carbonell WS, DeLay M, Jahangiri A, Park CC, Aghi MK. β1 Integrin targeting potentiates antiangiogenic therapy and inhibits the growth of bevacizumab-resistant glioblastoma. Cancer Res. 2013;73:3145–3154. doi: 10.1158/0008-5472.CAN-13-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Javadi S, Rostamizadeh K, Hejazi J, Parsa M, Fathi M. Curcumin mediated down-regulation of αVβ3 Integrin and up-regulation of pyruvate dehydrogenase kinase 4 (PDK4) in Erlotinib resistant SW480 colon cancer cells. Phytother Res. 2018;32:355–364. doi: 10.1002/ptr.5984. [DOI] [PubMed] [Google Scholar]
- 30.Fang RH, Aryal S, Hu CM, Zhang L. Quick synthesis of lipid-polymer hybrid nanoparticles with low polydispersity using a single-step sonication method. Langmuir. 2010;26:16958–16962. doi: 10.1021/la103576a. [DOI] [PubMed] [Google Scholar]
- 31.Lu Y, Zhong L, Jiang Z, Pan H, Zhang Y, Zhu G, Bai L, Tong R, Shi J, Duan X. Cationic micelle-based siRNA delivery for efficient colon cancer gene therapy. Nanoscale Res Lett. 2019;14:193. doi: 10.1186/s11671-019-2985-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang XZ, Dou S, Wang YC, Long HY, Xiong MH, Mao CQ, Yao YD, Wang J. Single-step assembly of cationic lipid-polymer hybrid nanoparticles for systemic delivery of siRNA. ACS Nano. 2012;6:4955–4965. doi: 10.1021/nn300500u. [DOI] [PubMed] [Google Scholar]
- 33.Pan B, Guo J, Liao Q, Zhao Y. β1 and β3 Integrins in breast, prostate and pancreatic cancer: a novel implication. Oncol Lett. 2018;15:5412–5416. doi: 10.3892/ol.2018.8076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nikolaou M, Pavlopoulou A, Georgakilas AG, Kyrodimos E. The challenge of drug resistance in cancer treatment: a current overview. Clin Exp Metastasis. 2018;35:309–318. doi: 10.1007/s10585-018-9903-0. [DOI] [PubMed] [Google Scholar]
- 35.Jahangiri A, Aghi MK, Carbonell WS. β1 integrin: critical path to antiangiogenic therapy resistance and beyond. Cancer Res. 2014;74:3–7. doi: 10.1158/0008-5472.CAN-13-1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stoeltzing O, Liu W, Reinmuth N, Fan F, Parry GC, Parikh AA, McCarty MF, Bucana CD, Mazar AP, Ellis LM. Inhibition of Integrin α5β1 function with a small peptide (ATN-161) plus continuous 5-FU infusion reduces colorectal liver metastases and improves survival in mice. Int J Cancer. 2003;104:496–503. doi: 10.1002/ijc.10958. [DOI] [PubMed] [Google Scholar]
- 37.DeLay M, Jahangiri A, Carbonell WS, Hu YL, Tsao S, Tom MW, Paquette J, Tokuyasu TA, Aghi MK. Microarray analysis verifies two distinct phenotypes of glioblastomas resistant to antiangiogenic therapy. Clin Cancer Res. 2012;18:2930–2942. doi: 10.1158/1078-0432.CCR-11-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mu X, Lu H, Fan L, Yan S, Hu K. Efficient delivery of therapeutic siRNA with nanoparticles induces apoptosis in prostate cancer cells. J Nanomater. 2018;2018 [Google Scholar]
- 39.Dong H, Yao L, Bi W, Wang F, Song W, Lv Y. Combination of survivin siRNA with neoadjuvant chemotherapy enhances apoptosis and reverses drug resistance in breast cancer MCF-7 cells. J Cancer Res Ther. 2015;11:717. doi: 10.4103/0973-1482.147764. [DOI] [PubMed] [Google Scholar]


