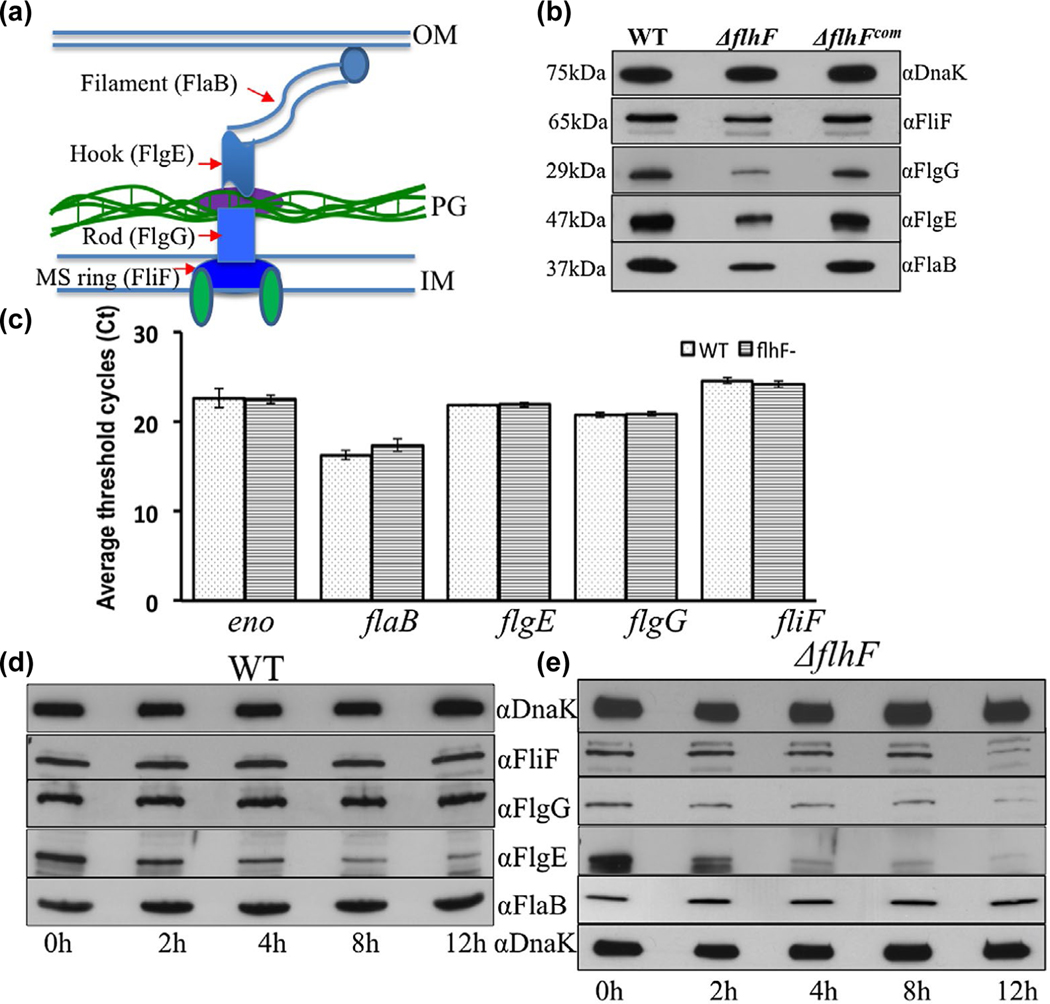
FIGURE 5.
Deletion of flhFBb affects the level and stability of flagellar proteins in B. burgdorferi. (a) A diagram illustrating the overall structure of PF. Red arrows denote four flagellar proteins that represent the MS-ring (FliF), the rod (FlgG), the hook (FlgE) and the filament (FlaB). (b) Detection of FliF, FlgG, FlgE and FlaB by immunoblotting analyses. Similar amounts of WT, ΔflhF and ΔflhFcom whole-cell lysates were subjected to SDS-PAGE and then probed with specific antibodies against these four proteins respectively. DnaK was used as a loading control. Immunoblots were developed using horseradish peroxidase secondary antibody with an ECL luminol substrate as previously described (Zhang et al., 2019). (c) Detection of four flagellar gene transcripts by qRT-PCR. The levels of flaB, flgE, flgG and fliF transcripts were measured by qRT-PCR as previously described (Sal et al., 2008; Sze et al., 2011). The transcript of enolase gene (eno) was used as an internal control to normalize the qPCR data. The results were expressed as the mean threshold cycles (CT) of triplicate samples. (d) & (e) Protein turnover assays. For this assay, protein translation was arrested by adding spectinomycin (100 μg/ml) to the cultures of WT and ΔflhF mutant, and then samples were collected at the indicated time points and subjected to immunoblotting. Of note, compared to the wild type, the loading amount of the flhF mutant was increased about 3-folds in order to detect these flagellar proteins during a period of 12 hr