Figure 3.
Cryo-EM 3D reconstruction of S protein at pH 6.5
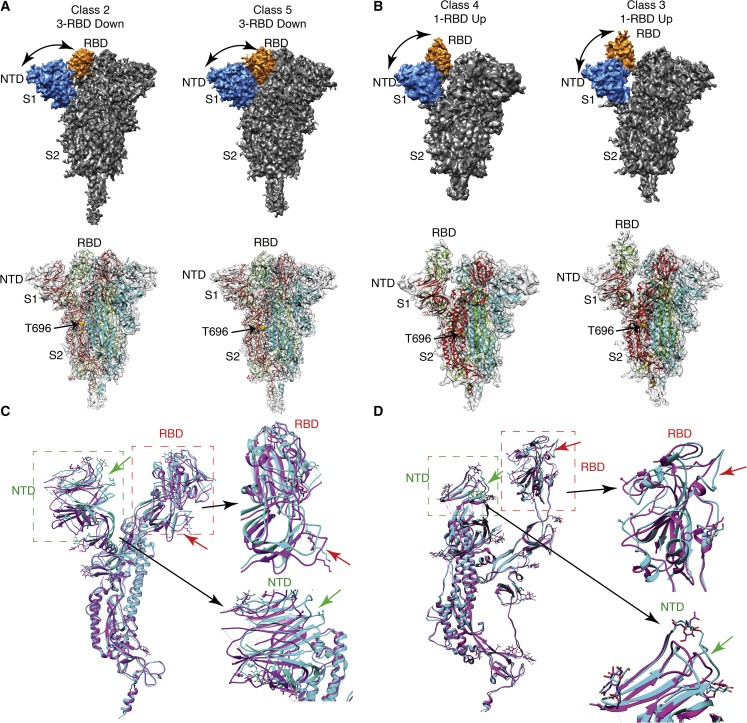
(A and B) Solid and transparent representation of cryo-EM 3D reconstruction of S protein at pH 6.5. The transparent representation of S protein is fitted with an atomic model calculated from 3D reconstructions of S protein and PDB: 6vyb (1-RBD up) and PDB: 6zwv (3-RBD down). Yellow spheres represent T696 at the boundary of S1 and S2 region. The RBD and NTD of the solid representation of the S protein are colored orange and blue, respectively. At pH 6.5, two 3-RBD down-closed conformations (A) and two 1-RBD up-open conformations (B) are observed. Distance between RBD and NTD is marked by black curved arrows.
(C) Comparison between the atomic models of two closed (all-RBD down conformation) states, class 2 (cyan) and class 5 (purple), indicates the movement of RBD and NTD. RBD and NTD are marked by red and green boxes, respectively. Black arrows show the enlarged view of RBD and NTD, where significant displacement is observed.
(D) Comparison between atomic models of two open states, class 4 (cyan) and class 3 (purple). Difference in NTD region is shown by green arrows and RBD region by red arrows.
See also Figure S4 and Tables S1–S3.