Fig. 1.
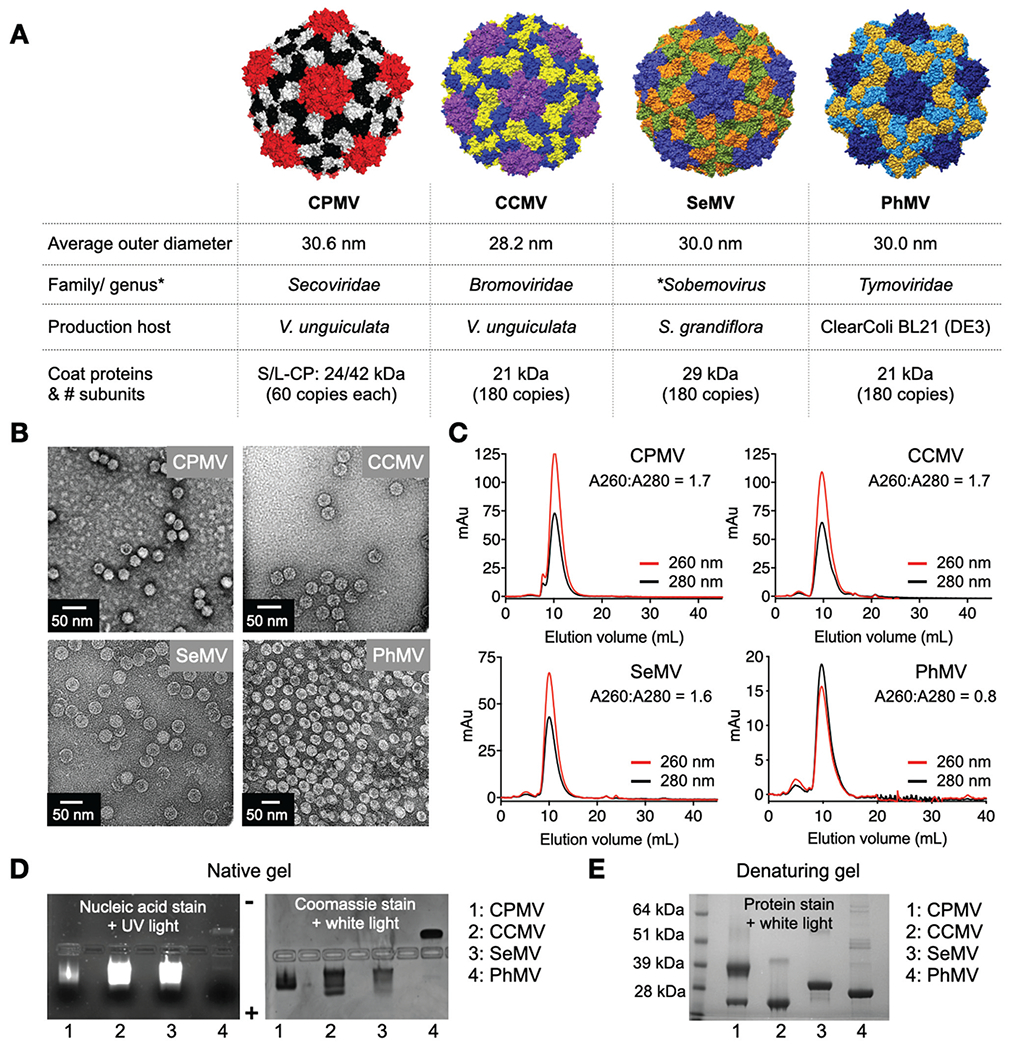
Plant viral nanoparticles. (A) Structure of CPMV, was created using Pymol (v2.3.4) using Protein Data Bank (PDB) entry 1NY7; CCMV, SeMV and PhMV space filling models were created on UCSF Chimera software 1.12 using PDB entries 1CWP, 1SMV and 1E57, respectively. Information on size is as reported on *VIPERdb. (B) TEM images of negatively stained virus nanoparticles. (C) SEC analysis shows characteristics elution profiles from a Superose 6 column; the A260 : A280 ratio indicates presence (~1.8) or absence (0.8) of nucleic acid. (D) Native agarose gel electrophoresis stained with GelRed® nucleic acid stain and Coomassie Brilliant Blue; gels were imaged under UV and white light. (E) Denaturing 4–12% Nu-PAGE gel stained with GelCode™ Blue Safe protein stain shows the coat proteins for the various virus preparations (*VIPERdb).32
