Abstract
Background
Members of the adhesion family of G protein-coupled receptors (GPCRs) have received attention for their roles in health and disease, including cancer. Over the past decade, several members of the family have been implicated in the pathogenesis of glioblastoma.
Methods
Here, we discuss the basic biology of adhesion GPCRs and review in detail specific members of the receptor family with known functions in glioblastoma. Finally, we discuss the potential use of adhesion GPCRs as novel treatment targets in neuro-oncology.
Keywords: adhesion GPCR, glioblastoma, G protein-coupled receptor
Glioblastoma
Glioma is the most common primary brain malignancy.1 Advances in genomics over the past decade have identified distinct driver mutations and transcriptional programs, which have led to re-classification of the glioma family. The most notable classifier in adult gliomas is neomorphic mutations in isocitrate dehydrogenase (IDH) 1 and 2. These mutations identify one type of glioma, predominantly seen in young adults, which generally has a more indolent course.2 IDH wild-type tumors, which include most glioblastomas (GBMs), are commonly associated with mutations in the TERT (telomerase) promoter and cause rapid neurologic decline and death.3 The Cancer Genome Atlas has used bulk transcriptome profiles to obtain gene expression signatures that further classify GBM into subtypes (classical, proneural, and mesenchymal).4 Similar subtypes can be derived by analyzing the DNA methylome of tumors.5 Nevertheless, GBM tumors display immense intratumoral heterogeneity and subtype-spanning plasticity.6–8 Regardless of their mutational status and transcriptome profile, gliomas are not curable by surgical excision due to their propensity to invade brain tissue.6,9 At the same time, gliomas, and GBM in particular, evade chemoradiotherapy through a variety of tumor cell-intrinsic and microenvironment-mediated mechanisms. Therapy resistance has been partly attributed to a cellular hierarchy dominated by stem-like cells, which are not only particularly adept at repairing DNA damage inflicted by chemoradiotherapy, but also capable of initiating tumor growth and generating all tumor lineages.10–17
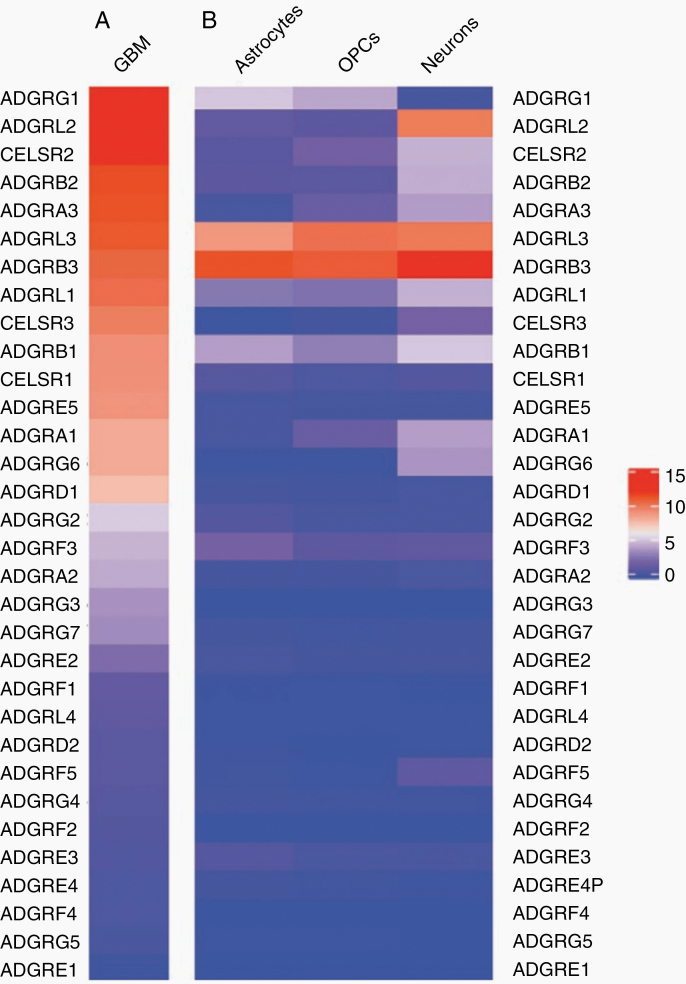
The fact that gliomas are almost universally lethal and evade radiotherapy, conventional chemotherapy, anti-angiogenic therapy, targeted therapies, and, so far, immunotherapy, highlights the need for identifying new treatment targets. In search of such new targets, we started studying adhesion G protein-coupled receptors (aGPCRs) in GBM several years ago. As Figure 1A illustrates, analysis of our previously published RNA-sequencing data from our patient-derived GBM cultures18 using R indicates that several aGPCRs are expressed by tumor cells. In contrast, several of the aGPCRs expressed in GBM are absent from normal brain tissue, as evidenced by single-cell RNA-sequencing data from normal brain tissue (Allen Brain Atlas; https://celltypes.brain-map.org/rnaseq/human_m1_10x; Figure 1B). This suggests that several aGPCRs are de novo expressed in GBM. As a result, here we propose that aGPCRs may offer appealing opportunities for novel therapies in glioma.
Figure 1.
Comparison of aGPCR transcript levels in GBM and normal brain. (A) Heatmap showing ranked log2(FPKM) aGPCR transcript levels from averaged RNA-sequencing data of 2 patient-derived IDH wild-type GBM cultures.18 The 2 cultures were transcriptionally subtyped as proneural and mesenchymal and were in culture for 5 passages before sequencing. (B) Heatmap of averaged astrocyte, oligodendrocyte precursor cell (OPC), and neuron transcript level values are from Allen Brain Atlas Human Multiple Cortex Areas SMART-seq data. The ranking of aGPCRs is identical to that in (A). Data represent averaged log2(CPM) values from layer 1–6 cortical astrocytes (n = 966), layer 1–6 cortical OPCs (n = 773), and excitatory and inhibitory neuronal clusters (n = 7382). The gene expression heatmaps were generated with R.
Classification and General Characteristics of adhesion G protein-coupled receptors
Adhesion GPCRs comprise 33 members in the human genome and represent the second largest family within the GPCR superfamily.19,20 According to recent classification systems, they are divided into 9 subfamilies, namely ADGRA, ADGRB, ADGRC, ADGRD, ADGRE, ADGRF, ADGRG, ADGRL, and ADGRV, although new taxonomies have recently emerged.21 In this review, we will primarily refer to the aGPCRs by their original names.
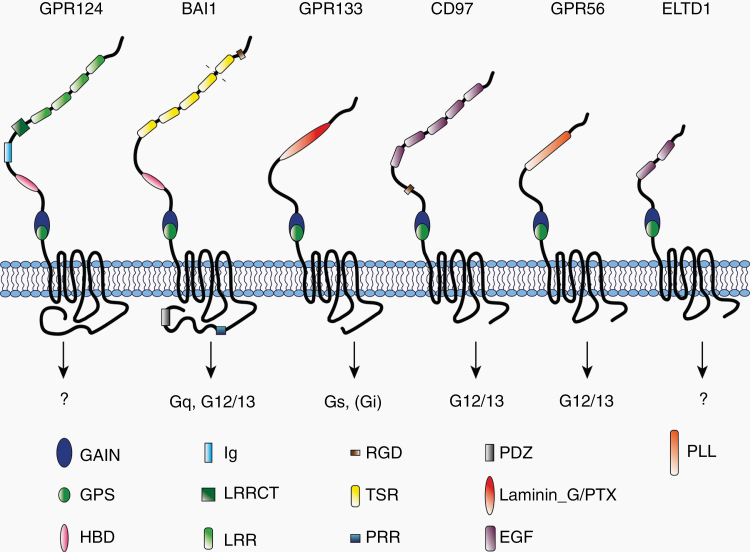
Like all GPCRs, members of the aGPCR family are structurally defined by 7 conserved α-helical transmembrane loops (7-TM domain), an intracellular C-terminus, and an extracellular N-terminus. What distinguishes aGPCRs from other GPCRs, however, is their long N-terminus, which varies in length and functional subdomain composition based on the receptor subtype (Figure 2). These functional domains have been shown to convey cell–cell or extracellular matrix (ECM) interactions, suggesting that these receptors have a dual role as cell adhesion and signaling proteins.20 All aGPCRs, with the exception of GPR123, possess a conserved GPCR autoproteolysis-inducing (GAIN) domain in the N-terminus that catalyzes cleavage at a GPCR proteolysis site (GPS) to generate an N-terminal and a C-terminal fragment (NTF and CTF, respectively).22 The processes following proteolysis have not been fully elucidated, but there is evidence that the NTF and CTF may remain non-covalently bound to each other in the secretory pathway and dissociate after being trafficked to the plasma membrane. Immediately distal to the GPS lies an endogenous agonist sequence, named the Stachel sequence, which is responsible for activating canonical signaling. Soluble peptides derived from this tethered agonist sequence have been used to experimentally modulate aGPCR function.23–31
Figure 2.
Functional domains and G protein coupling of aGPCRs implicated in GBM. The schematic shows structures and functional domains at the NTF of GPR124, BAI1, GPR133, CD97, GPR56, CELSR1, and ELTD1. G protein coupling is indicated by arrows. To date, G protein coupling of GPR124 and ELTD1 has not been documented. GAIN, GPCR autoproteolysis-inducing domain; GPS, GPCR proteolysis site; HBD, hormone-binding domain; Ig, immunoglobulin domain; LRRCT, leucine-rich repeat C-terminal domain; LRR, leucine-rich repeat; RGD, Arg-Gly-Asp motif; TSR, thrombospondin type 1 repeat; PRR, proline-rich region; laminin_G/PTX, laminin_G/pentraxin; EGF, epidermal growth factor domain; PLL, pentraxin/laminin/neurexin/sex-hormone-binding-globulin-like domain.
To date, there are numerous publications that provide data on aGPCR canonical signaling via G proteins. Coupling to Gαs, Gαi, Gα12/13, or Gαq proteins has been shown for many of the receptors.20,26,29,32,33 G protein-independent non-canonical signaling has also been reported for aGPCRs. The most prominent examples are the BAI family of aGPCRs and GPR124 (ADGRA2), which are involved in Rac-1-mediated signaling34–36 and Wnt pathways,37,38 respectively.
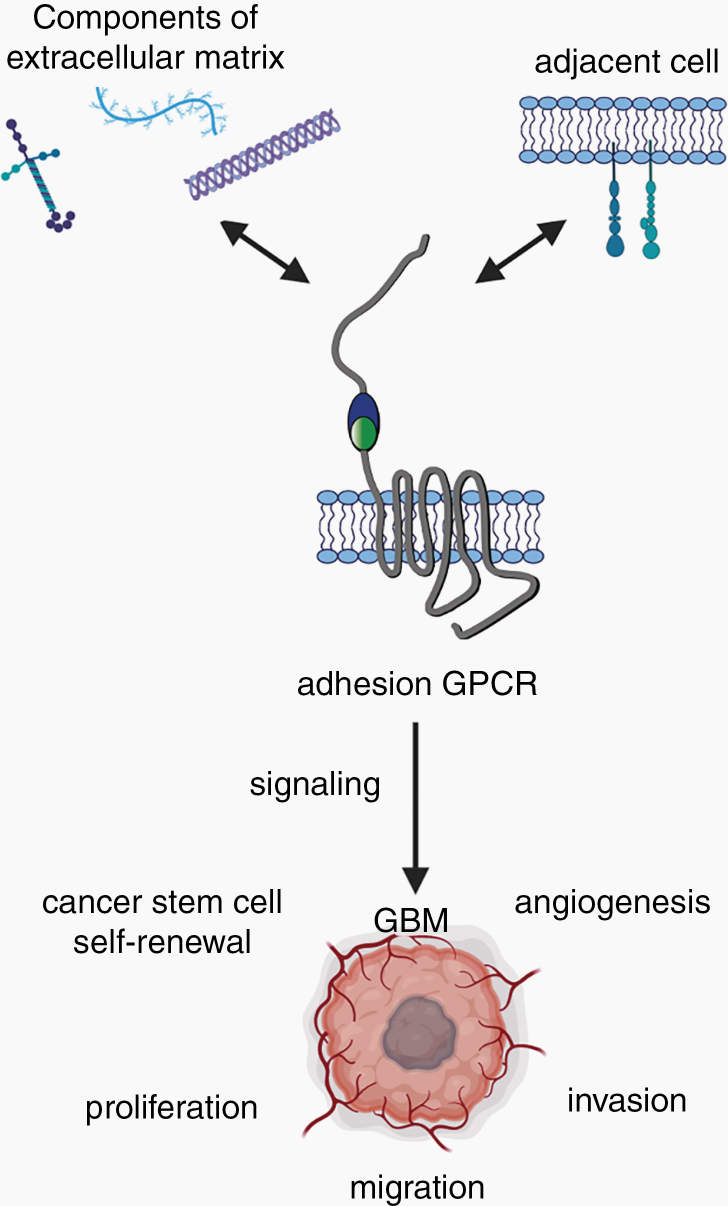
aGPCRs play pivotal roles in physiological cellular processes, such as establishing cell shape and polarity, mediating cell adhesion and migration, and transmitting mechanical stimuli.39–44 At the organismal level, aGPCRs have been implicated in the immune response, endocrine and nervous system function, and tumorigenesis.20,21,45,46 Moreover, aGPCRs are involved in brain development, establishment of the blood–brain barrier (BBB) and regulation of brain angiogenesis, and may contribute to the stemness of GBM stem cells (Zhou 2014, Kuhnert 2000, Kuhnert 2010, Cullen 2011, Nishimori 1997, Bayin 2016).37,40,47,48,49 Most importantly for the purposes of this review, several members of the aGPCR family have been implicated in glioma biology. Thus, we will focus our review on specific aGPCRs relevant for GBM and analyze their function within the context of tumor cell migration, brain invasion, cellular proliferation, stem cell self-renewal, and angiogenesis (Figure 3).
Figure 3.
Impact of aGPCRs on GBM biology. aGPCRs bind ligands from the ECM or the plasma membrane of adjacent cells, thereby conveying cell–cell or cell–matrix interactions. Binding of a ligand results in receptor activation and either canonical signaling via G proteins or non-canonical signaling. This leads to changes in cellular processes, such as cell proliferation, migration, angiogenesis, and invasion, directly impacting GBM biology.
Specific Adhesion G Protein-Coupled Receptors in GBM
Several aGPCRs have been implicated in GBM tumorigenesis. Here, we will focus on some of the specific aGPCRs that our analysis indicates are upregulated in our patient-derived GBM cultures (Figure 1A) and that are most prominent in GBM research. These include GPR124, BAI1, GPR133, CD97, EMR2, GPR56, and ELTD1. Some aGPCRs, which demonstrate similar expression patterns, for example, members of the family of cadherin EGF LAG seven-pass G-type receptors, are purposely left out of the review due to a lack of relevant literature implicating them in GBM. For the aGPCRs reviewed here, we will discuss the structural and functional properties of each receptor and provide up-to-date information regarding their implication in the oncogenic process, with a particular focus on GBM. Since several of these aGPCRs are expressed in both tumor and endothelial cells, we will review their function in both cellular contexts where appropriate.
GPR124 (ADGRA2)
GPR124 is an orphan receptor, also known as TEM5 (tumor endothelial marker 5). According to recent taxonomy arrangements, it belongs to subfamily III of aGPCRs.20 GPR124’s serine/threonine-rich N-terminus is characterized by leucine-rich repeats, a leucine-rich repeat C-terminal domain, an immunoglobulin (Ig) domain, and a hormone-binding domain (HBD; Figure 2). Thrombin-induced shedding cleaves the receptor at the HBD into an NTF and CTF.50 The association/dissociation of NTF and CTF is protein disulfide isomerase (PDI)-dependent.50 Thrombin-induced cleavage exposes an RGD motif that mediates cell adhesion by binding integrins.50,51 A more recent study also suggests that GPR124 is involved in cell adhesion via the interaction with the Rho guanine exchange factors Elmo/Dock and intersectin through its C-terminus.52
Numerous publications highlighted GPR124’s involvement in Wnt signaling in the brain endothelium and the receptor’s key role in angiogenesis and development of the brain vasculature.37,38,47,53 Both in vitro and in vivo studies suggested that GPR124 serves as a co-activator for canonical Wnt signaling via Frizzled and Lrp receptors and Wnt7a/7b.37,38 Additionally, GPR124 interacts with Reck, a GPI-anchored matrix metalloprotease (MMP) inhibitor, to build a signaling complex at the level of the plasma membrane, thereby contributing to Wnt signaling in brain angiogenesis and BBB formation.54–60
The impact of GPR124 on adult forebrain angiogenesis and the establishment of the BBB were further investigated by producing an inducible conditional knockout in mouse endothelial cells. Endothelial GPR124 deficiency led to BBB disruption, increased tumor hemorrhage, and decreased survival in a GBM mouse model.61 Interestingly, the proliferation capacity of cultured GBM cells was significantly reduced by both tumor cell-specific overexpression and knockdown of the receptor.62 Transcript levels of GPR124 are not detected in RNA-Seq datasets from the Allen Brain Atlas in normal brain cells (Figure 1B), while it is moderately expressed in patient-derived GBM cell lines (Figure 1A).
Collectively, most data suggest that GPR124 is mainly expressed in tumor vasculature, is upregulated in GBM, and plays a major role in Wnt signaling, an important pathway for brain angiogenesis and BBB formation. GPR124 may merit further investigation as a target in GBM, because the suppression of its pro-angiogenic function is predicted to inhibit tumor growth and progression.
BAI1 (ADGRB1)
Subfamily VII of the aGPCR family comprises the 3 brain-specific angiogenesis inhibitor (BAI) genes: BAI1, BAI2, and BAI3.21,63 Like all BAIs, BAI1 harbors thrombospondin type 1 repeat domains and an HBD within its N-terminus, as well as a C-terminal PDZ domain.63 Of all 3 BAIs, only BAI1 contains an N-terminal RDG motif, an MMP-14 site, and a C-terminal proline-rich region (Figure 2). BAI1 is involved in both canonical G protein signaling via Gαq and Gα12/1326 and non-canonical signaling leading to Rho pathway activation, phosphorylation of ERK, and β-arrestin binding.64 Recently, peptides derived from BAI1’s endogenous Stachel sequence were designed and used to activate the receptor in neurons, where it binds Neuroligin-1, a cell-adhesion molecule found at synapses.65 The Stachel peptide-induced activation resulted in Rac-1 activation and synapse development, highlighting the role of BAI1 in synaptogenesis.65
The first evidence for BAI1’s involvement in GBM was given in 1997, when Nishimori et al.48 found that the receptor is expressed in normal brain cells, but its transcript is significantly decreased in established GBM cell lines. Several other studies agreed with those findings, observing repeated detection of BAI1 in normal glial cells at both the transcript and protein levels, while failing to detect its presence in GBM cells.66 Consistent with these findings is the observation that BAI1 expression decreases with rising malignancy grades in glioma tumors.67 RNA-seq data from our laboratory show only moderate BAI1 expression in patient-derived GBM cells in vitro (Figure 1A), while it is one of the top 5 detected transcripts in normal brain cells from the Allen Brain Atlas (Figure 1B). A recent study suggests that BAI1 is epigenetically downregulated in GBM by hypermethylation of its promoter region.68 In this study, the following evidence suggested that methyl-CpG-binding domain protein 2 (MBD2), an epigenetic regulator of gene expression, is responsible for the downregulation of BAI1 in GBM: (1) treatment of GBM cells with 5-Aza-2′-deoxycytidine, a DNA demethylating agent, restores BAI1 expression; (2) chromatin immunoprecipitation shows enrichment of MBD2 at the BAI1 promoter region; and (3) shRNA-mediated knockdown of MDB2 leads to the re-activation of BAI1 expression in glioma cells.
BAI1 can be cleaved at its GPS, autoproteolytically, resulting in a 120 kDa NTF (Vasculostatin-120), or at its MMP-14 site, resulting in a 40 kDa NTF (Vasculostatin-40). Both cleavage products have been shown to contribute to physiological processes within the brain. Vasculostatin-120 decreases intracranial glioma growth in vivo, while both Vasculostatin-120 and Vasculostatin-40 were suggested to increase anti-angiogenic and anti-tumorigenic effects in normal brain and GBM.69–71 In orthotopic xenografts implanted in rats, Vasculostatin-120 reduces intracranial growth of malignant gliomas and tumor vascular density, even upon a pro-angiogenic stimulus.71 In endothelial cells, the anti-angiogenic effect was suggested to be dependent on the surface molecule CD36.71 Likewise, the anti-angiogenic and anti-tumorigenic effects of full-length BAI1 were shown in xenograft models in vivo,72 independent of P53 expression within the tumor.66,73 Taken together, these findings suggest a tumor-suppressive role for BAI1 in GBM. The identification of agents that restore expression of BAI1 could potentially serve as a therapeutic tool for the treatment of GBM.
GPR133 (ADGRD1)
According to recent taxonomy arrangements, GPR133 belongs to subfamily V of aGPCRs.20 In addition to the GAIN domain and the GPS, GPR133’s N-terminal ectodomain contains a laminin G/pentraxin (LMN/PTX) domain (Figure 2). As shown in other aGPCRs, the C-terminal sequence immediately following the cleavage site within the GPS represents the endogenous tethered Stachel agonist, which is responsible for activating GPR133 as confirmed by mutational studies.23 Deleting the NTF of GPR133 leads to increased receptor activity.23 Initial insights into GPR133 canonical signaling and G protein binding were produced by a few recent studies. Upon GPR133 heterologous expression in Cos-7 and HEK293T cells, cAMP levels increase significantly, an effect that is eliminated with Gαs subunit knockdown.74–76 This indicates that the GPR133 receptor couples with the Gαs subunit upon activation. GPR133 signaling is increased by administering soluble peptides derived from the endogenous Stachel sequence to Cos-7 cells expressing the receptor.23
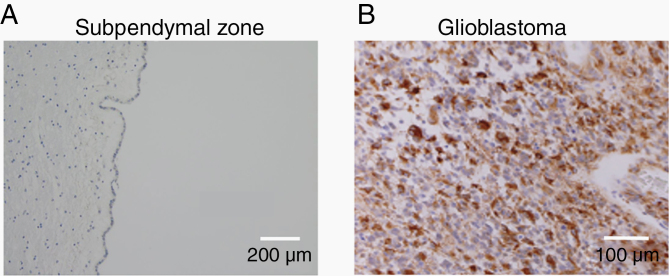
GPR133, whose ligands remain unknown, was recently shown to be necessary for tumor growth in GBM.49,77 Knockdown of GPR133 by shRNA results in reduced cell proliferation and tumorsphere formation in vitro. Furthermore, GPR133 knockdown impairs orthotopic tumor xenograft initiation in vivo.49 RNA-Seq data from GBM cells show GPR133 transcript expression (Figure 1A), while it is not detected in neurons, astrocytes, and oligodendrocyte precursor cells (OPCs; Figure 1B). Frenster et al.78 used immunohistochemistry to show that GPR133 is essentially de novo expressed in GBM, because it is absent from normal brain tissue (Figure 4). Importantly, GPR133 expression was detected in both IDH wild-type and IDH mutant tumors.78 Furthermore, the same study suggested a positive correlation between GPR133 expression and the WHO grade of gliomas, raising the possibility that GPR133 is a marker of anaplasia in the glioma family.
Figure 4.
GPR133 is de novo expressed in GBM. Representative micrographs of GPR133 immunohistochemistry78 from the subependymal zone of non-neoplastic cadaveric brain (A) and an IDH wild-type GBM (B). The subependymal zone around the brain ventricular system contains progenitor cells that may represent the putative cell of origin for glioma.
GPR133 is enriched in the most hypoxic regions of GBM, also known as areas of pseudopalisading necrosis. This phenomenon is mediated by transcriptional upregulation of GPR133 in hypoxia via direct binding of hypoxia-inducible factor 1α (HIF-1α) to its promoter.49 The finding suggests that GPR133 is not only a necessary component of GBM growth, but it may also mediate the tumor’s cellular response to hypoxia. Collectively, these data suggest that GPR133 merits further consideration as a potential target in GBM. It is therefore necessary to identify inhibitory ligands, neutralizing antibodies or small molecule inhibitors of GPR133, or to engineer antibody-drug conjugates (ADCs) as therapeutics for GBM treatment.
CD97 (ADGRE5) and EMR2 (ADGRE2)
CD97 is an aGPCR from subfamily II, consisting of 5 total EGF-TM7 receptors, aptly named for a series of epidermal growth factor (EGF) repeats found in the N-terminal ectodomain79 (Figure 2). In humans, the longest CD97 receptor isoform contains 5 EGF domains (EGF [1–5]), while 2 shorter isoforms result from alternative splicing and contain 3 and 4 EGF domains (EGF [1,2,5] and EGF [1,2,3,5], respectively). Autoproteolytic cleavage is observed at the GPS, which resides within the characteristic GAIN domain, and is essential for proper receptor trafficking and function.79
In addition to EGF domains, the NTF contains an RGD motif and several N-glycosylation sites. It also mediates interaction with ligands, which include CD55, chondroitin sulfate, CD90, and multiple integrins, such as α5β1.80 CD55 and chondroitin sulfate are both membrane-associated macromolecules expressed by immune cells and demonstrate cell-to-cell interaction with CD97.81 Integrin α5β1, also known as the fibronectin receptor, is a transmembrane protein that interacts with ECM components and is involved in the angiogenic process.82 This ligand is an attractive drug target as it is upregulated in a series of solid tumors, including GBM.83 The CTF of CD97 consists of the 7-TM domain and a short intracellular domain that has been shown to interact with the Gα12/13 subunit during receptor activation. This is corroborated by RhoA activation upon ectopic CD97 expression.84 Nevertheless, CD97 has also been shown to couple with other G protein signaling pathways, such as Gβγ85 and Gz, where it dampens cAMP levels.86
CD97 has been widely studied within the context of the immune system, where the receptor facilitates migration and adhesion of leukocytes to sites of inflammation87 and has been implicated in the regulation of acute myeloid leukemia stem cells.88 New research shows an aberrant expression of the receptor in a multitude of solid tumors, including GBM.89 CD97 is observed to impart both an invasive and a migratory phenotype on GBM cells.79 CD97 expression is absent from normal brain tissue, but 2 splice variants (EGF [1,2,5] and EGF [1,2,3,5]) are highly expressed in GBM, especially in the mesenchymal and classical subtypes.90 Since many of the ligands that interact with CD97 do so via the EGF domains, it has been established that the different isoforms demonstrate unique ligand-binding capacities. For example, only the largest isoform recognizes chondroitin sulfate,81 while the smallest isoform is most likely to recognize CD55.91 This introduces new questions of ligand–receptor binding and how it impacts receptor processing and signaling. For example, do different ligands activate/inhibit different signaling pathways? Does the availability of a particular ligand shift isoform expression? Can interruption of a ligand–receptor interaction shift ligand preference?
EMR2 (ADGRE2) belongs to the same subfamily as CD97 and shares many similarities, including several EGF domains along its extracellular domain.20 High EMR2 expression has been associated with low-grade gliomas and the mesenchymal subtype of GBM.92 Ivan et al.92 found a correlation between EMR2 and the PI3K pathway, observing that both were upregulated in GBM following therapy with bevacizumab, a monoclonal antibody against VEGF-A occasionally used to treat GBM. A study has also observed that EMR2 contributes to an invasive phenotype and correlates with poor survival in GBM patients.93 Similar to CD97, EMR2 demonstrates binding to chondroitin sulfate.94
Overall, CD97 and EMR2 may represent exciting drug targets due to their high expression levels in multiple solid tumors, including GBM. Existing evidence suggests that the 2 receptors promote cellular migration and invasion, a phenotype of GBM cells linked to their aggressive behavior and poor patient prognosis. Nevertheless, growing evidence suggests that CD97 also regulates other processes, such as maintenance of the stem cell hierarchy and facilitation of cellular adhesion. Further research is needed to elucidate the function of CD97 and the impact that targeting the receptor may have on cancer progression, including in GBM.
GPR56 (ADGRG1)
GPR56 belongs to the aGPCR subfamily VIII20 and is arguably the most broadly studied aGPCR within an oncological context. The receptor contains a pentraxin/laminin/neurexin/sex-hormone-binding-globulin-like (PLL) domain within its N-terminus (Figure 2), shown to be essential for ligand binding.95 Alternative splicing of GPR56 generates multiple isoforms, one of which is termed splice variant 4 (S4) and completely lacks the PLL domain.95 The receptor also contains a series of N- and O-linked glycosylation sites along its extracellular domain.96
Ligands of GPR56 include the ECM components collagen-III and transglutaminase-2 (TG2).97 Both of these proteins have been found to facilitate NTF dissociation after receptor cleavage. The binding of TG2 to the NTF of GPR56 causes the receptor–ECM complex to be internalized and degraded by the cell.98 GPR56, therefore, may play a role in ECM remodeling (reviewed in Ref. 99), which is an essential aspect of GBM cell migration and invasion. The receptor also binds heparin, a glycosaminoglycan that interacts with other ECM components.100 GPR56 activation has been observed in both a Stachel-dependent and Stachel-independent manner.101 The receptor is known to couple with the Gα12/13 subunit to activate the Rho signaling pathway.102 Non-canonical signaling by GPR56 includes modulation of the PI3K/AKT103 and β-catenin96 pathways. Though GPR56 has mainly been implicated in oncogenic processes such as cellular adhesion, migration, and ECM remodeling, the receptor also seems to promote an anti-angiogenic response by reducing VEGF secretion.104
RNA-seq data from our laboratory show that GPR56 is the most abundantly expressed aGPCR in patient-derived GBM cells (Figure 1A), whereas single-cell SMART-seq data from the Allen Brain Atlas suggest that GPR56 expression is low in neurons and moderate in astrocytes and OPCs from normal brain tissue (Figure 1B). From the developmental point of view, GPR56 plays a crucial role in brain development, neural progenitor migration, and differentiation in the oligodendrocyte lineage and has been linked with polymicrogyria.40,105–109 Immunohistochemistry against GPR56 reveals its increased abundance within GBM tissue compared to normal brain tissue.96 The aGPCR seems to be particularly concentrated at membrane extensions (such as filopodia) and co-localizes with actin filaments at focal adhesion points within GBM cells in vitro.96 Moreno et al.110 found that GPR56 is primarily expressed in proneural and classical GBM subtypes and determined that the receptor inhibits the transition of these subtypes into the mesenchymal phenotype. While Shashidhar et al.96 found that GPR56 activates several oncogenic signaling cascades, including the NF-κB pathway, Moreno et al.110 proposed that the receptor actually inhibits the NF-κB pathway and linked its high expression in GBM tissue with better survival outcome and less radioresistance. Additional studies have found that GPR56 both suppresses and promotes cancer progression, further highlighting the conflicting role GPR56 plays in tumor biology.
Though the receptor’s exact role in GBM biology remains controversial, GPR56 serves as an important bridge facilitating connections between the extracellular and intracellular environment. Its impact on cancer progression is likely context-dependent and tissue-specific. Targeting of GPR56 with both small molecule inhibitors and monoclonal antibodies has been shown to modulate receptor signaling,31,97 but its overall function in GBM remains unclear.
ELTD1 (ADGRL4)
ELTD1 (EGF, latrophilin, and 7 transmembrane domain-containing protein 1) is an aGPCR within Group I, which additionally contains 3 latrophilin receptors.20 Favara et al.111 comprehensively reviewed this receptor in 2014, detailing its structure and functions. The extracellular domain contains both an EGF domain and an EGF Ca2+-binding domain112 (Figure 2). One variant of the receptor is truncated at the C-terminal end.111 Currently, ELTD1 remains an orphan receptor and little is known about its posttranslational processing and signaling. In our bulk RNA-seq data, we find only modest ELTD1 expression in patient-derived GBM cells; however, we have included it in this review due to extensive available literature implicating the receptor in GBM biology and associated angiogenesis (Figure 1A).
ELTD1 has emerged as an angiogenic biomarker, co-regulated with other angiogenic factors, such as VEGF, NOTCH1, and DLL4.113,114ELTD1 is transcriptionally upregulated in blood vessels of high-grade glioma tumors compared to vessels from low-grade gliomas and from nonmalignant control tissue. Immunohistochemical analysis confirmed the expression of ELTD1 in vascular-associated cells.113 Li et al. demonstrated that ELTD1 acts by stimulating the JAK/STAT signaling pathway115 and increases the expression of HIF-1α,116 the transcription factor that serves as a master regulator of the hypoxic response and driver of vascularization.117 Since ELTD1 serves as an angiogenic marker, studies have attempted to target the receptor in the hopes of halting tumor vascularization and ultimately progression. Immunohistochemical approaches showed that ELTD1 co-localizes with the VEGF receptor (VEGFR) in mouse tumor tissue.118 When neutralizing antibodies were used against ELTD1, VEGFR protein levels decreased. Similarly, ELTD1 protein levels decreased upon treatment with a VEGFR neutralizing antibody.118 Furthermore, in vivo administration of a monoclonal antibody targeting ELTD1 reduced GBM tumor volume and microvessel density compared to an untreated control.114 These studies highlight the potential use of anti-ELTD1 neutralizing antibodies for anti-angiogenic therapy of GBM tumors.114,118,119
Studies have also demonstrated ELTD1 expression in GBM tissue itself, especially in the mesenchymal subtype, where receptor levels correlate with GBM progression and poor prognosis.115 Functionally, ELTD1 overexpression in established GBM cell lines promotes proliferation, migration, and invasion.116 Conversely, ELTD1 knockdown reduces GBM cellular viability,120 proliferation, and invasion capacity in vitro and decreases tumorigenesis in vivo, effects that are overcome by HIF-1α overexpression.116
Ultimately, neovascularization at the site of the tumor enables rapid GBM progression. Initial experiments targeting ELTD1 in mice showed promising results, effectively reducing GBM growth and vascularization. Nevertheless, clinical trials testing the effects of the anti-angiogenic drug bevacizumab on GBM outcome showed little effect on overall patient survival. It is possible that combined targeting of several angiogenic markers, such as VEGFR and ELTD1, could help reduce treatment resistance.
Adhesion G Protein-Coupled Receptors as Potential Therapeutic Targets for Glioblastoma Treatment
Currently, no therapies targeting aGPCRs are approved or in clinical trials, although the unique features of aGPCRs show promising opportunities.121 In general, aGPCRs may be attractive therapeutic targets for various reasons: (1) with the exception of BAI1, aGPCRs discussed in this review are upregulated in GBM or tumor-associated vasculature compared to normal brain tissue/vasculature; (2) aGPCRs are plasma membrane proteins, which in principle makes them more accessible to BBB-permeant therapeutics relative to intracellular targets; and (3) their long extracellular N-termini with distinct domains could serve as binding sites for specific biologics. The latter feature is most relevant if the NTF and CTF of the aGPCR are associated at the plasma membrane, even after autoproteolytic cleavage of the receptor.
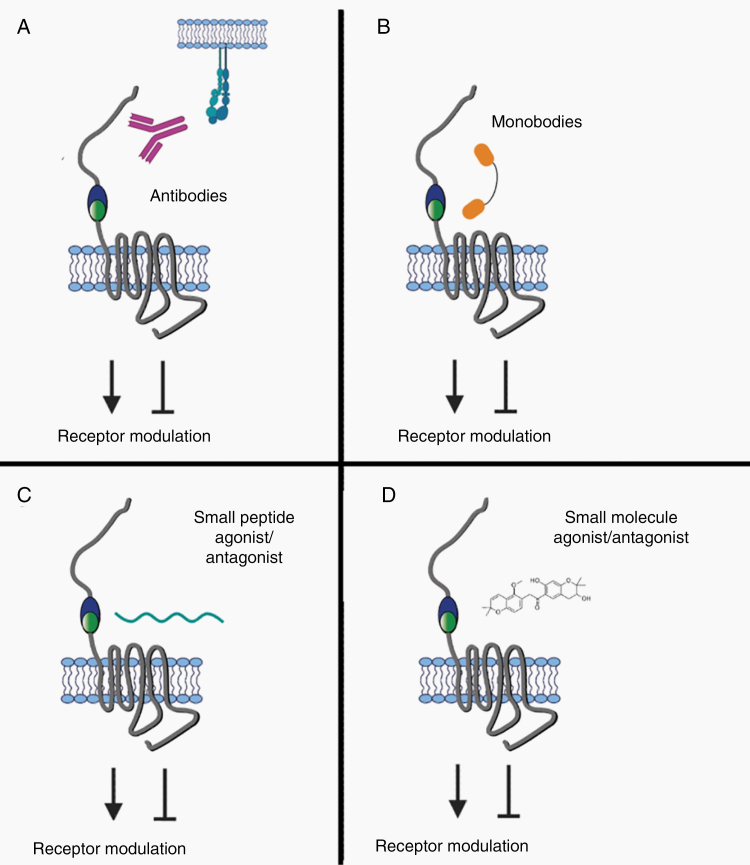
Targeting aGPCRs in GBM with antibodies could be achieved via different strategies (Figure 5). Therapeutic neutralizing antibodies may either block ligand-binding sites or otherwise prevent receptor activation and signaling. Such action could therefore inhibit aGPCR-related processes that primarily promote tumor growth and progression, such as cellular proliferation, migration, invasion, or angiogenesis. Both in vivo and in vitro experiments have demonstrated successful targeting of aGPCRs with neutralizing antibodies. In the case of ELTD1, receptor targeting with a monoclonal antibody led to reduced GBM tumor volume and vascularization in a glioma mouse model.114 A CD97 antibody demonstrated target specificity, cellular internalization, and safe pharmacokinetics in mice and even reduced the inflammatory response in an arthritic mouse model compared to those treated with a control,122 while an antibody recognizing EMR2 facilitated leukocyte migration.39 These examples show that antibodies targeting the extracellular domains of aGPCRs can have a range of impacts on receptor function. Another use of antibodies could be the targeted transport of cytotoxic therapeutics to tumor cells using ADCs. Since many of the aGPCRs appear to have de novo expression in GBM, they could be ideal targets for ADC approaches. No studies to date have generated ADCs targeting aGPCRs.
Figure 5.
Approaches to modulating aGPCRs as targets toward novel therapeutics. (A) Antibodies that interfere with receptor–ligand interactions can modulate receptor function and signaling. Alternatively, antibodies that target the receptor and lead to internalization may be used to deliver cytotoxic cargo in ADC approaches. (B) Similar to antibodies, monobodies can modulate receptor activity or deliver cytotoxic agents upon internalization, but with the added advantage of smaller size. (C and D) Small peptide agonists and antagonists (C), derived from endogenous Stachel sequences, and small molecules (D) also represent viable approaches to modulating aGPCR signaling.
As an alternative to antibodies, monobodies are a novel biologic platform for targeting aGPCRs (Figure 5). Monobodies are synthetic binding proteins based on a fibronectin type III domain with an immunoglobulin fold, but without any disulfide bonds.123 While they can be engineered to have antibody-like target specificity, their smaller size may afford easier permeation through the BBB when GBM therapies are considered. Different monobodies against GPR56 were shown to both increase and decrease GPR56-mediated signaling in HEK239T cells and hence modulate the receptor in vitro.101 By targeting the PLL domain of GPR56 with monobodies, Salzman et al.124 were able to disrupt receptor interaction with TG2. The development of monobodies against other aGPCRs implicated in GBM and their testing in vivo would serve as an advanced tool in discovering new therapeutic options.
Targeting the ligands of aGPCRs may also be a viable approach (Figure 5). For example, a small molecule inhibitor (TTGM 5826) against TG2, a GPR56 ligand, has shown success in vitro by reducing the growth of breast cancer and GBM cells.125 Other promising candidates are CD55 or integrin α5β1, which serve as CD97 ligands. Knockdown of CD55 attenuated growth of prostate cancer cells126 and an integrin α5β1 inhibitor was successfully used to attenuate glioma growth.127 In fact, multiple integrin α5β1-selective biologics are currently in clinical trials (as reviewed in Refs 128,129). These examples help demonstrate the utility of targeting ligands toward modulating aGPCR function.
As discussed previously, peptides derived from the Stachel sequence have been successfully used as aGPCR agonists, modulating signaling and receptor function in vitro21,23–31 (Figure 5). In principle, such peptides could be mutated to inhibit aGPCR activation. However, their hydrophobic character, low solubility, and low potency currently limit possible clinical applications.
The conventional pharmacologic strategy to modulate aGPCR signaling in GBM involves small molecules, typically identified via high-throughput screening (Figure 5). To date, GPR56 and GPR114 have been successfully inhibited by the small molecule antagonist dihydromunduletone in vitro.130 A small molecule partial agonist of GPR56 was found to mediate Gα13 activation.31 Moreover, decylubiquinone, which modulates the ROS/P53/BAI1 signaling pathway and increases BAI1 expression, reduces breast cancer growth and metastasis in a mouse model. Together, these studies suggest that the use of small molecule drugs to modulate aGPCR signaling and function is a promising approach in the treatment of cancer.131
In conclusion, we review compelling evidence that several aGPCRs are de novo expressed in GBM and serve primarily pro-tumorigenic roles, with the exception of BAI1, whose functional profile suggests tumor-suppressive properties. Specific aGPCRs have demonstrated direct involvement in a series of oncogenic processes, including cellular migration and invasion (CD97), stem cell self-renewal (GPR133), ECM remodeling (GPR124, GPR56, CD97), and vascularization (GPR124, BAI1, ELTD1). Given their expression profile, presence on the plasma membrane of tumor cells, potential “druggability,” and essential roles in tumorigenesis, we propose that aGPCRs represent putative novel targets in GBM. With this therapeutic potential in mind, we review exisiting data on small molecules and biologics that modulate aGPCR function and suggest opportunities for therapy development.
Funding
This study was supported by National Institute of Neurological Disorders and Stroke R01 NS102665 and New York State Stem Cell Science IIRP C32595GG to D.G.P. D.G.P. was also supported by NIH/National Institute of Biomedical Imaging and Bioengineering R01 EB028774, NYU Grossman School of Medicine, and Deutche Forschungsgemeinschaft (DFG) (German Research Foundation) FOR2149. G.S. was supported by a DFG postdoctoral fellowship (STE 2843/1-1). N.R.B. was supported by a T32 training grant (T32GM136542) to the Cell Biology Training Program at NYU Grossman School of Medicine.
Conflict of interest statement. D.G.P. and NYU Grossman School of Medicine hold a patent titled “Method for treating high grade glioma” on the use of GPR133 as a treatment target in glioma. D.G.P. has received consultant fees from Tocagen, Synaptive Medical, Monteris, and Robeaute.
Authorship Statement. G.S., N.R.B., and D.G.P.: Manuscript preparation.
References
- 1. Dolecek TA, Propp JM, Stroup NE, Kruchko C. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2005–2009. Neuro Oncol. 2012;14(suppl 5):v1–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cohen AL, Holmen SL, Colman H. IDH1 and IDH2 mutations in gliomas. Curr Neurol Neurosci Rep. 2013;13(5):345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pekmezci M, Rice T, Molinaro AM, et al. . Adult infiltrating gliomas with WHO 2016 integrated diagnosis: additional prognostic roles of ATRX and TERT. Acta Neuropathol. 2017;133(6):1001–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Verhaak RG, Hoadley KA, Purdom E, et al. ; Cancer Genome Atlas Research Network . Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ma H, Zhao C, Zhao Z, et al. . Specific glioblastoma multiforme prognostic-subtype distinctions based on DNA methylation patterns. Cancer Gene Ther. 2020;27(9):702–714. [DOI] [PubMed] [Google Scholar]
- 6. Omuro A, DeAngelis LM. Glioblastoma and other malignant gliomas: a clinical review. JAMA. 2013;310(17):1842–1850. [DOI] [PubMed] [Google Scholar]
- 7. Weller M, et al. Glioma. Nat Rev Dis Primers. 2015;1:15017. doi: 10.1038/nrdp.2015.17 [DOI] [PubMed] [Google Scholar]
- 8. Behnan J, Finocchiaro G, Hanna G. The landscape of the mesenchymal signature in brain tumours. Brain. 2019;142(4):847–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li C, Wang S, Yan JL, et al. . Characterizing tumor invasiveness of glioblastoma using multiparametric magnetic resonance imaging. J Neurosurg. 2019;132(5):1–8. doi: 10.3171/2018.12.JNS182926 [DOI] [PubMed] [Google Scholar]
- 10. Lathia JD, Mack SC, Mulkearns-Hubert EE, Valentim CL, Rich JN. Cancer stem cells in glioblastoma. Genes Dev. 2015;29(12):1203–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Alcantara Llaguno SR, Parada LF. Cell of origin of glioma: biological and clinical implications. Br J Cancer. 2016;115(12):1445–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Singh SK, Hawkins C, Clarke ID, et al. . Identification of human brain tumour initiating cells. Nature. 2004;432(7015):396–401. [DOI] [PubMed] [Google Scholar]
- 13. Bayin NS, Modrek AS, Placantonakis DG. Glioblastoma stem cells: molecular characteristics and therapeutic implications. World J Stem Cells. 2014;6(2):230–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen J, Li Y, Yu TS, et al. . A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488(7412):522–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang R, Chadalavada K, Wilshire J, et al. . Glioblastoma stem-like cells give rise to tumour endothelium. Nature. 2010;468(7325):829–833. [DOI] [PubMed] [Google Scholar]
- 16. Bao S, Wu Q, McLendon RE, et al. . Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444(7120):756–760. [DOI] [PubMed] [Google Scholar]
- 17. Lan X, Jörg DJ, Cavalli FMG, et al. . Fate mapping of human glioblastoma reveals an invariant stem cell hierarchy. Nature. 2017;549(7671):227–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bayin NS, Frenster JD, Sen R, et al. . Notch signaling regulates metabolic heterogeneity in glioblastoma stem cells. Oncotarget. 2017;8(39):64932–64953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Krishnan A, Nijmeijer S, de Graaf C, Schiöth HB. Classification, nomenclature, and structural aspects of adhesion GPCRs. Handb Exp Pharmacol. 2016;234:15–41. [DOI] [PubMed] [Google Scholar]
- 20. Hamann J, Aust G, Araç D, et al. . International Union of Basic and Clinical Pharmacology. XCIV. Adhesion G protein-coupled receptors. Pharmacol Rev. 2015;67(2):338–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Scholz N, Langenhan T, Schöneberg T. Revisiting the classification of adhesion GPCRs. Ann N Y Acad Sci. 2019;1456(1):80–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Araç D, Boucard AA, Bolliger MF, et al. . A novel evolutionarily conserved domain of cell-adhesion GPCRs mediates autoproteolysis. EMBO J. 2012;31(6):1364–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liebscher I, Schön J, Petersen SC, et al. . A tethered agonist within the ectodomain activates the adhesion G protein-coupled receptors GPR126 and GPR133. Cell Rep. 2014;9(6):2018–2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Demberg LM, Rothemund S, Schöneberg T, Liebscher I. Identification of the tethered peptide agonist of the adhesion G protein-coupled receptor GPR64/ADGRG2. Biochem Biophys Res Commun. 2015;464(3):743–747. [DOI] [PubMed] [Google Scholar]
- 25. Müller A, Winkler J, Fiedler F, et al. . Oriented cell division in the C. elegans embryo is coordinated by g-protein signaling dependent on the adhesion GPCR LAT-1. PLoS Genet. 2015;11(10):e1005624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kishore A, Purcell RH, Nassiri-Toosi Z, Hall RA. Stalk-dependent and Stalk-independent signaling by the adhesion G protein-coupled receptors GPR56 (ADGRG1) and BAI1 (ADGRB1). J Biol Chem. 2016;291(7):3385–3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wilde C, Fischer L, Lede V, et al. . The constitutive activity of the adhesion GPCR GPR114/ADGRG5 is mediated by its tethered agonist. FASEB J. 2016;30(2):666–673. [DOI] [PubMed] [Google Scholar]
- 28. Demberg LM, Winkler J, Wilde C, et al. . Activation of adhesion G protein-coupled receptors: agonist specificity of Stachel sequence-derived peptides. J Biol Chem. 2017;292(11):4383–4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Purcell RH, Toro C, Gahl WA, Hall RA. A disease-associated mutation in the adhesion GPCR BAI2 (ADGRB2) increases receptor signaling activity. Hum Mutat. 2017;38(12):1751–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Scholz N, Guan C, Nieberler M, et al. . Mechano-dependent signaling by Latrophilin/CIRL quenches cAMP in proprioceptive neurons. Elife. 2017;6:e28360. doi: 10.7554/eLife.28360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Stoveken HM, Larsen SD, Smrcka AV, Tall GG. Gedunin- and khivorin-derivatives are small-molecule partial agonists for adhesion g protein-coupled receptors GPR56/ADGRG1 and GPR114/ADGRG5. Mol Pharmacol. 2018;93(5):477–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Purcell RH, Hall RA. Adhesion G protein-coupled receptors as drug targets. Annu Rev Pharmacol Toxicol. 2018;58:429–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mathiasen S, Palmisano T, Perry NA, et al. . G12/13 is activated by acute tethered agonist exposure in the adhesion GPCR ADGRL3. Nat Chem Biol. 2020;16(12):1343–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Park D, Tosello-Trampont AC, Elliott MR, et al. . BAI1 is an engulfment receptor for apoptotic cells upstream of the ELMO/Dock180/Rac module. Nature. 2007;450(7168):430–434. [DOI] [PubMed] [Google Scholar]
- 35. Duman JG, Tzeng CP, Tu YK, et al. . The adhesion-GPCR BAI1 regulates synaptogenesis by controlling the recruitment of the Par3/Tiam1 polarity complex to synaptic sites. J Neurosci. 2013;33(16):6964–6978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Weng Z, Situ C, Lin L, Wu Z, Zhu J, Zhang R. Structure of BAI1/ELMO2 complex reveals an action mechanism of adhesion GPCRs via ELMO family scaffolds. Nat Commun. 2019;10(1):51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhou Y, Nathans J. Gpr124 controls CNS angiogenesis and blood-brain barrier integrity by promoting ligand-specific canonical wnt signaling. Dev Cell. 2014;31(2):248–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Posokhova E, Shukla A, Seaman S, et al. . GPR124 functions as a WNT7-specific coactivator of canonical β-catenin signaling. Cell Rep. 2015;10(2):123–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yona S, Lin HH, Dri P, et al. . Ligation of the adhesion-GPCR EMR2 regulates human neutrophil function. FASEB J. 2008;22(3):741–751. [DOI] [PubMed] [Google Scholar]
- 40. Koirala S, Jin Z, Piao X, Corfas G. GPR56-regulated granule cell adhesion is essential for rostral cerebellar development. J Neurosci. 2009;29(23):7439–7449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Scholz N, Gehring J, Guan C, et al. . The adhesion GPCR latrophilin/CIRL shapes mechanosensation. Cell Rep. 2015;11(6):866–874. [DOI] [PubMed] [Google Scholar]
- 42. Nijmeijer S, Vischer HF, Leurs R. Adhesion GPCRs in immunology. Biochem Pharmacol. 2016;114:88–102. [DOI] [PubMed] [Google Scholar]
- 43. Lin HH, Hsiao CC, Pabst C, Hébert J, Schöneberg T, Hamann J. Adhesion GPCRs in regulating immune responses and inflammation. Adv Immunol. 2017;136:163–201. [DOI] [PubMed] [Google Scholar]
- 44. Langenhan T. Adhesion G protein-coupled receptors—candidate metabotropic mechanosensors and novel drug targets. Basic Clin Pharmacol Toxicol. 2020;126(suppl 6):5–16. [DOI] [PubMed] [Google Scholar]
- 45. Lawrence PA, Struhl G, Casal J. Planar cell polarity: one or two pathways? Nat Rev Genet. 2007;8(7):555–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Aust G, Zhu D, Van Meir EG, Xu L. Adhesion GPCRs in tumorigenesis. Handb Exp Pharmacol. 2016;234:369–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kuhnert F, Mancuso MR, Shamloo A, et al. . Essential regulation of CNS angiogenesis by the orphan G protein-coupled receptor GPR124. Science. 2010;330(6006):985–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nishimori H, Shiratsuchi T, Urano T, et al. . A novel brain-specific p53-target gene, BAI1, containing thrombospondin type 1 repeats inhibits experimental angiogenesis. Oncogene. 1997;15(18):2145–2150. [DOI] [PubMed] [Google Scholar]
- 49. Bayin NS, Frenster JD, Kane JR, et al. . GPR133 (ADGRD1), an adhesion G-protein-coupled receptor, is necessary for glioblastoma growth. Oncogenesis. 2016;5(10):e263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vallon M, Aubele P, Janssen KP, Essler M. Thrombin-induced shedding of tumour endothelial marker 5 and exposure of its RGD motif are regulated by cell-surface protein disulfide-isomerase. Biochem J. 2012;441(3):937–944. [DOI] [PubMed] [Google Scholar]
- 51. Ruoslahti E. RGD and other recognition sequences for integrins. Annu Rev Cell Dev Biol. 1996;12:697–715. [DOI] [PubMed] [Google Scholar]
- 52. Hernández-Vásquez MN, Adame-García SR, Hamoud N, et al. . Cell adhesion controlled by adhesion G protein-coupled receptor GPR124/ADGRA2 is mediated by a protein complex comprising intersectins and Elmo-Dock. J Biol Chem. 2017;292(29):12178–12191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Weinsheimer S, Brettman AD, Pawlikowska L, et al. . G protein-coupled receptor 124 (GPR124) gene polymorphisms and risk of brain arteriovenous malformation. Transl Stroke Res. 2012;3(4):418–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Vanhollebeke B, Stone OA, Bostaille N, et al. . Tip cell-specific requirement for an atypical Gpr124- and Reck-dependent Wnt/β-catenin pathway during brain angiogenesis. Elife. 2015;4:e06489. doi: 10.7554/eLife.06489 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bostaille N, Gauquier A, Twyffels L, Vanhollebeke B. Molecular insights into Adgra2/Gpr124 and Reck intracellular trafficking. Biol Open. 2016;5(12):1874–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Alok A, Lei Z, Jagannathan NS, et al. . Wnt proteins synergize to activate β-catenin signaling. J Cell Sci. 2017;130(9):1532–1544. [DOI] [PubMed] [Google Scholar]
- 57. Cho C, Smallwood PM, Nathans J. Reck and Gpr124 are essential receptor cofactors for Wnt7a/Wnt7b-specific signaling in mammalian CNS angiogenesis and blood-brain barrier regulation. Neuron. 2017;95(5):1056–1073.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Eubelen M, Bostaille N, Cabochette P, et al. . A molecular mechanism for Wnt ligand-specific signaling. Science (New York, N.Y.). 2018;361(6403):eaat1178. doi: 10.1126/science.aat1178 [DOI] [PubMed] [Google Scholar]
- 59. Vallon M, Yuki K, Nguyen TD, et al. . A RECK-WNT7 receptor-ligand interaction enables isoform-specific regulation of Wnt bioavailability. Cell Rep. 2018;25(2):339–349.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Cho C, Wang Y, Smallwood PM, Williams J. & Nathans J. Molecular determinants in Frizzled, Reck, and Wnt7a for ligand-specific signaling in neurovascular development. Elife. 2019;8:e47300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Chang J, Mancuso MR, Maier C, et al. . Gpr124 is essential for blood-brain barrier integrity in central nervous system disease. Nat Med. 2017;23(4):450–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Cherry AE, Vicente JJ, Xu C, et al. . GPR124 regulates microtubule assembly, mitotic progression, and glioblastoma cell proliferation. Glia. 2019;67(8):1558–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Cork SM, Van Meir EG. Emerging roles for the BAI1 protein family in the regulation of phagocytosis, synaptogenesis, neurovasculature, and tumor development. J Mol Med (Berl). 2011;89(8):743–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Stephenson JR, Paavola KJ, Schaefer SA, Kaur B, Van Meir EG, Hall RA. Brain-specific angiogenesis inhibitor-1 signaling, regulation, and enrichment in the postsynaptic density. J Biol Chem. 2013;288(31):22248–22256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Tu YK, Duman JG, Tolias KF. The adhesion-GPCR BAI1 promotes excitatory synaptogenesis by coordinating bidirectional trans-synaptic signaling. J Neurosci. 2018;38(39):8388–8406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kaur B, Brat DJ, Calkins CC, Van Meir EG. Brain angiogenesis inhibitor 1 is differentially expressed in normal brain and glioblastoma independently of p53 expression. Am J Pathol. 2003;162(1):19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wang W, Da R, Wang M, et al. . Expression of brain-specific angiogenesis inhibitor 1 is inversely correlated with pathological grade, angiogenesis and peritumoral brain edema in human astrocytomas. Oncol Lett. 2013;5(5):1513–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zhu D, Hunter SB, Vertino PM, Van Meir EG. Overexpression of MBD2 in glioblastoma maintains epigenetic silencing and inhibits the antiangiogenic function of the tumor suppressor gene BAI1. Cancer Res. 2011;71(17):5859–5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kaur B, Brat DJ, Devi NS, Van Meir EG. Vasculostatin, a proteolytic fragment of brain angiogenesis inhibitor 1, is an antiangiogenic and antitumorigenic factor. Oncogene. 2005;24(22):3632–3642. [DOI] [PubMed] [Google Scholar]
- 70. Cork SM, Kaur B, Devi NS, et al. . A proprotein convertase/MMP-14 proteolytic cascade releases a novel 40 kDa vasculostatin from tumor suppressor BAI1. Oncogene. 2012;31(50):5144–5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kaur B, Cork SM, Sandberg EM, et al. . Vasculostatin inhibits intracranial glioma growth and negatively regulates in vivo angiogenesis through a CD36-dependent mechanism. Cancer Res. 2009;69(3):1212–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kang X, Xiao X, Harata M, et al. . Antiangiogenic activity of BAI1 in vivo: implications for gene therapy of human glioblastomas. Cancer Gene Ther. 2006;13(4):385–392. [DOI] [PubMed] [Google Scholar]
- 73. Berger B, Capper D, Lemke D, et al. . Defective p53 antiangiogenic signaling in glioblastoma. Neuro Oncol. 2010;12(9):894–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Bohnekamp J, Schöneberg T. Cell adhesion receptor GPR133 couples to Gs protein. J Biol Chem. 2011;286(49):41912–41916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Gupte J, Swaminath G, Danao J, Tian H, Li Y, Wu X. Signaling property study of adhesion G-protein-coupled receptors. FEBS Lett. 2012;586(8):1214–1219. [DOI] [PubMed] [Google Scholar]
- 76. Fischer L, Wilde C, Schöneberg T, Liebscher I. Functional relevance of naturally occurring mutations in adhesion G protein-coupled receptor ADGRD1 (GPR133). BMC Genomics. 2016;17(1):609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Frenster JD, Inocencio JF, Xu Z, et al. . GPR133 promotes glioblastoma growth in hypoxia. Neurosurgery. 2017;64(suppl 1):177–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Frenster JD, Kader M, Kamen S, et al. . Expression profiling of the adhesion G protein-coupled receptor GPR133 (ADGRD1) in glioma subtypes. Neurooncol Adv. 2020;2(1):vdaa053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Safaee M, Clark AJ, Ivan ME, et al. . CD97 is a multifunctional leukocyte receptor with distinct roles in human cancers (Review). Int J Oncol. 2013;43(5):1343–1350. [DOI] [PubMed] [Google Scholar]
- 80. Wang T, Ward Y, Tian L, et al. . CD97, an adhesion receptor on inflammatory cells, stimulates angiogenesis through binding integrin counterreceptors on endothelial cells. Blood. 2005;105(7):2836–2844. [DOI] [PubMed] [Google Scholar]
- 81. Kwakkenbos MJ, Pouwels W, Matmati M, et al. . Expression of the largest CD97 and EMR2 isoforms on leukocytes facilitates a specific interaction with chondroitin sulfate on B cells. J Leukoc Biol. 2005;77(1):112–119. [DOI] [PubMed] [Google Scholar]
- 82. Boudreau NJ, Varner JA. The homeobox transcription factor Hox D3 promotes integrin alpha5beta1 expression and function during angiogenesis. J Biol Chem. 2004;279(6):4862–4868. [DOI] [PubMed] [Google Scholar]
- 83. Schaffner F, Ray AM, Dontenwill M. Integrin α5β1, the fibronectin receptor, as a pertinent therapeutic target in solid tumors. Cancers (Basel). 2013;5(1):27–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Ward Y, Lake R, Yin JJ, et al. . LPA receptor heterodimerizes with CD97 to amplify LPA-initiated RHO-dependent signaling and invasion in prostate cancer cells. Cancer Res. 2011;71(23):7301–7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Hilbig D, Sittig D, Hoffmann F, et al. . Mechano-dependent phosphorylation of the PDZ-binding motif of CD97/ADGRE5 modulates cellular detachment. Cell Rep. 2018;24(8):1986–1995. [DOI] [PubMed] [Google Scholar]
- 86. Bhudia N, Desai S, King N, et al. . Author correction: G protein-coupling of adhesion GPCRs ADGRE2/EMR2 and ADGRE5/CD97, and activation of G protein signalling by an anti-EMR2 antibody. Sci Rep. 2020;10(1):5097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Leemans JC, te Velde AA, Florquin S, et al. . The epidermal growth factor-seven transmembrane (EGF-TM7) receptor CD97 is required for neutrophil migration and host defense. J Immunol. 2004;172(2):1125–1131. [DOI] [PubMed] [Google Scholar]
- 88. Martin GH, Roy N, Chakraborty S, et al. . CD97 is a critical regulator of acute myeloid leukemia stem cell function. J Exp Med. 2019;216(10):2362–2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Steinert M, Wobus M, Boltze C, et al. . Expression and regulation of CD97 in colorectal carcinoma cell lines and tumor tissues. Am J Pathol. 2002;161(5):1657–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Safaee M, Fakurnejad S, Bloch O, et al. . Proportional upregulation of CD97 isoforms in glioblastoma and glioblastoma-derived brain tumor initiating cells. PLoS One. 2015;10(2):e0111532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Hamann J, Stortelers C, Kiss-Toth E, et al. . Characterization of the CD55 (DAF)-binding site on the seven-span transmembrane receptor CD97. Eur J Immunol. 1998;28:1701–1707. [DOI] [PubMed] [Google Scholar]
- 92. Ivan ME, Safaee M, Oh T, et al. . Epidermal growth factor-like module containing mucin-like hormone receptor 2 expression in gliomas. J Neurooncol. 2015;121(1):53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Rutkowski MJ, Sughrue ME, Kane AJ, Kim JM, Bloch O, Parsa AT. Epidermal growth factor module-containing mucin-like receptor 2 is a newly identified adhesion G protein-coupled receptor associated with poor overall survival and an invasive phenotype in glioblastoma. J Neurooncol. 2011;105(2):165–171. [DOI] [PubMed] [Google Scholar]
- 94. Stacey M, Chang GW, Davies JQ, et al. . The epidermal growth factor-like domains of the human EMR2 receptor mediate cell attachment through chondroitin sulfate glycosaminoglycans. Blood. 2003;102(8):2916–2924. [DOI] [PubMed] [Google Scholar]
- 95. Salzman GS, Ackerman SD, Ding C, et al. . Structural basis for regulation of GPR56/ADGRG1 by its alternatively spliced extracellular domains. Neuron. 2016;91(6):1292–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Shashidhar S, Lorente G, Nagavarapu U, et al. . GPR56 is a GPCR that is overexpressed in gliomas and functions in tumor cell adhesion. Oncogene. 2005;24(10):1673–1682. [DOI] [PubMed] [Google Scholar]
- 97. Xu L, Begum S, Hearn JD, Hynes RO. GPR56, an atypical G protein-coupled receptor, binds tissue transglutaminase, TG2, and inhibits melanoma tumor growth and metastasis. Proc Natl Acad Sci U S A. 2006;103(24):9023–9028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Yang L, Chen G, Mohanty S, et al. . GPR56 regulates VEGF production and angiogenesis during melanoma progression. Cancer Res. 2011;71(16):5558–5568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Mehta P, Piao X. Adhesion G-protein coupled receptors and extracellular matrix proteins: roles in myelination and glial cell development. Dev Dyn. 2017;246(4):275–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Chiang NY, Chang GW, Huang YS, et al. . Heparin interacts with the adhesion GPCR GPR56, reduces receptor shedding, and promotes cell adhesion and motility. J Cell Sci. 2016;129(11):2156–2169. [DOI] [PubMed] [Google Scholar]
- 101. Salzman GS, Zhang S, Gupta A, Koide A, Koide S, Araç D. Stachel-independent modulation of GPR56/ADGRG1 signaling by synthetic ligands directed to its extracellular region. Proc Natl Acad Sci U S A. 2017;114(38):10095–10100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Chiang NY, Peng YM, Juang HH, et al. . GPR56/ADGRG1 activation promotes melanoma cell migration via NTF dissociation and CTF-mediated Gα12/13/RhoA signaling. J Invest Dermatol. 2017;137(3):727–736. [DOI] [PubMed] [Google Scholar]
- 103. Ji B, Feng Y, Sun Y, et al. . GPR56 promotes proliferation of colorectal cancer cells and enhances metastasis via epithelial-mesenchymal transition through PI3K/AKT signaling activation. Oncol Rep. 2018;40(4):1885–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Yang L, Friedland S, Corson N, Xu L. GPR56 inhibits melanoma growth by internalizing and degrading its ligand TG2. Cancer Res. 2014;74(4):1022–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Giera S, Deng Y, Luo R, et al. . The adhesion G protein-coupled receptor GPR56 is a cell-autonomous regulator of oligodendrocyte development. Nat Commun. 2015;6:6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Iguchi T, Sakata K, Yoshizaki K, Tago K, Mizuno N, Itoh H. Orphan G protein-coupled receptor GPR56 regulates neural progenitor cell migration via a G alpha 12/13 and Rho pathway. J Biol Chem. 2008;283(21):14469–14478. [DOI] [PubMed] [Google Scholar]
- 107. Li S, Jin Z, Koirala S, et al. . GPR56 regulates pial basement membrane integrity and cortical lamination. J Neurosci. 2008;28(22):5817–5826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Jin Z, Tietjen I, Bu L, et al. . Disease-associated mutations affect GPR56 protein trafficking and cell surface expression. Hum Mol Genet. 2007;16(16):1972–1985. [DOI] [PubMed] [Google Scholar]
- 109. Cauley ES, Hamed A, Mohamed IN, et al. . Overlap of polymicrogyria, hydrocephalus, and Joubert syndrome in a family with novel truncating mutations in ADGRG1/GPR56 and KIAA0556. Neurogenetics. 2019;20(2):91–98. [DOI] [PubMed] [Google Scholar]
- 110. Moreno M, Pedrosa L, Paré L, et al. . GPR56/ADGRG1 inhibits mesenchymal differentiation and radioresistance in glioblastoma. Cell Rep. 2017;21(8):2183–2197. [DOI] [PubMed] [Google Scholar]
- 111. Favara DM, Banham AH, Harris AL. A review of ELTD1, a pro-angiogenic adhesion GPCR. Biochem Soc Trans. 2014;42(6):1658–1664. [DOI] [PubMed] [Google Scholar]
- 112. Nechiporuk T, Urness LD, Keating MT. ETL, a novel seven-transmembrane receptor that is developmentally regulated in the heart. ETL is a member of the secretin family and belongs to the epidermal growth factor-seven-transmembrane subfamily. J Biol Chem. 2001;276(6):4150–4157. [DOI] [PubMed] [Google Scholar]
- 113. Dieterich LC, Mellberg S, Langenkamp E, et al. . Transcriptional profiling of human glioblastoma vessels indicates a key role of VEGF-A and TGFβ2 in vascular abnormalization. J Pathol. 2012;228(3):378–390. [DOI] [PubMed] [Google Scholar]
- 114. Zalles M, Smith N, Ziegler J, et al. . Optimized monoclonal antibody treatment against ELTD1 for GBM in a G55 xenograft mouse model. J Cell Mol Med. 2020;24(2):1738–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Towner RA, Jensen RL, Colman H, et al. . ELTD1, a potential new biomarker for gliomas. Neurosurgery. 2013;72(1):77–90; discussion 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Li J, Shen J, Wang Z, et al. . ELTD1 facilitates glioma proliferation, migration and invasion by activating JAK/STAT3/HIF-1α signaling axis. Sci Rep. 2019;9:13904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Rey S, Semenza GL. Hypoxia-inducible factor-1-dependent mechanisms of vascularization and vascular remodelling. Cardiovasc Res. 2010;86(2):236–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Ziegler J, Zalles M, Smith N, et al. . Targeting ELTD1, an angiogenesis marker for glioblastoma (GBM), also affects VEGFR2: molecular-targeted MRI assessment. Am J Nucl Med Mol Imaging. 2019;9(1):93–109. [PMC free article] [PubMed] [Google Scholar]
- 119. Ziegler J, Pody R, Coutinho de Souza P, et al. . ELTD1, an effective anti-angiogenic target for gliomas: preclinical assessment in mouse GL261 and human G55 xenograft glioma models. Neuro Oncol. 2017;19(2):175–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Serban F, Daianu O, Tataranu LG, et al. . Silencing of epidermal growth factor, latrophilin and seven transmembrane domain-containing protein 1 (ELTD1) via siRNA-induced cell death in glioblastoma. J Immunoassay Immunochem. 2017;38(1):21–33. [DOI] [PubMed] [Google Scholar]
- 121. Bassilana F, Nash M, Ludwig MG. Adhesion G protein-coupled receptors: opportunities for drug discovery. Nat Rev Drug Discov. 2019;18(11):869–884. [DOI] [PubMed] [Google Scholar]
- 122. de Groot DM, Vogel G, Dulos J, et al. . Therapeutic antibody targeting of CD97 in experimental arthritis: the role of antigen expression, shedding, and internalization on the pharmacokinetics of anti-CD97 monoclonal antibody 1B2. J Immunol. 2009;183(6):4127–4134. [DOI] [PubMed] [Google Scholar]
- 123. Sha F, Salzman G, Gupta A, Koide S. Monobodies and other synthetic binding proteins for expanding protein science. Protein Sci. 2017;26(5):910–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Salzman GS, Zhang S, Fernandez CG, Araç D, Koide S. Specific and direct modulation of the interaction between adhesion GPCR GPR56/ADGRG1 and tissue transglutaminase 2 using synthetic ligands. Sci Rep. 2020;10(1):16912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Katt WP, Blobel NJ, Komarova S, Antonyak MA, Nakano I, Cerione RA. A small molecule regulator of tissue transglutaminase conformation inhibits the malignant phenotype of cancer cells. Oncotarget. 2018;9(76):34379–34397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Loberg RD, Day LL, Dunn R, Kalikin LM, Pienta KJ. Inhibition of decay-accelerating factor (CD55) attenuates prostate cancer growth and survival in vivo. Neoplasia. 2006;8(1):69–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Färber K, Synowitz M, Zahn G, et al. . An alpha5beta1 integrin inhibitor attenuates glioma growth. Mol Cell Neurosci. 2008;39(4):579–585. [DOI] [PubMed] [Google Scholar]
- 128. Hou J, Yan D, Liu Y, Huang P, Cui H. The roles of integrin α5β1 in human cancer. Onco Targets Ther. 2020;13:13329–13344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Rocha LA, Learmonth DA, Sousa RA, Salgado AJ. αvβ3 and α5β1 integrin-specific ligands: from tumor angiogenesis inhibitors to vascularization promoters in regenerative medicine? Biotechnol Adv. 2018;36(1):208–227. [DOI] [PubMed] [Google Scholar]
- 130. Stoveken HM, Bahr LL, Anders MW, Wojtovich AP, Smrcka AV, Tall GG. Dihydromunduletone is a small-molecule selective adhesion G protein-coupled receptor antagonist. Mol Pharmacol. 2016;90(3):214–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Cao J, Liu X, Yang Y, et al. . Decylubiquinone suppresses breast cancer growth and metastasis by inhibiting angiogenesis via the ROS/p53/ BAI1 signaling pathway. Angiogenesis. 2020;23(3):325–338. [DOI] [PubMed] [Google Scholar]